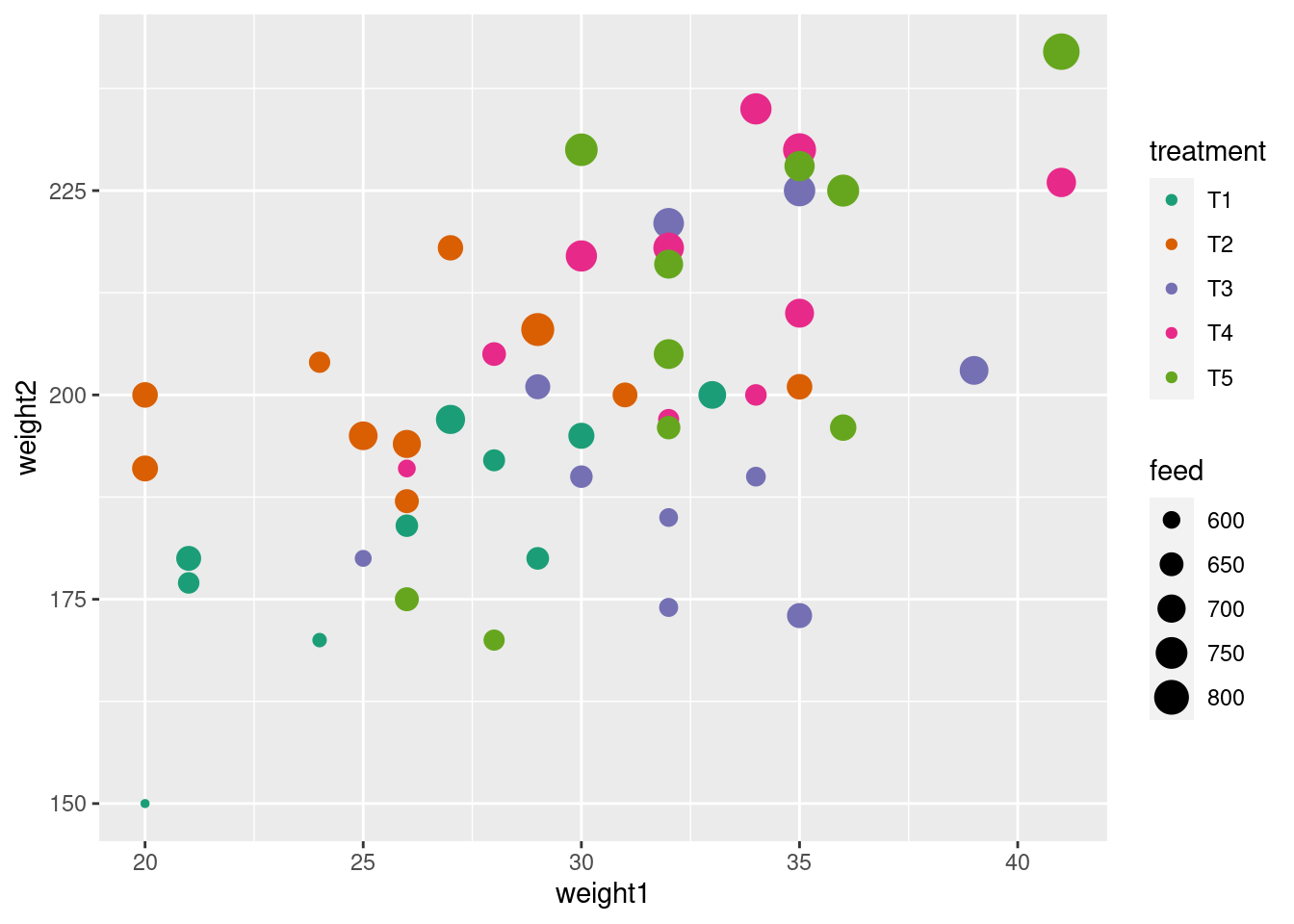
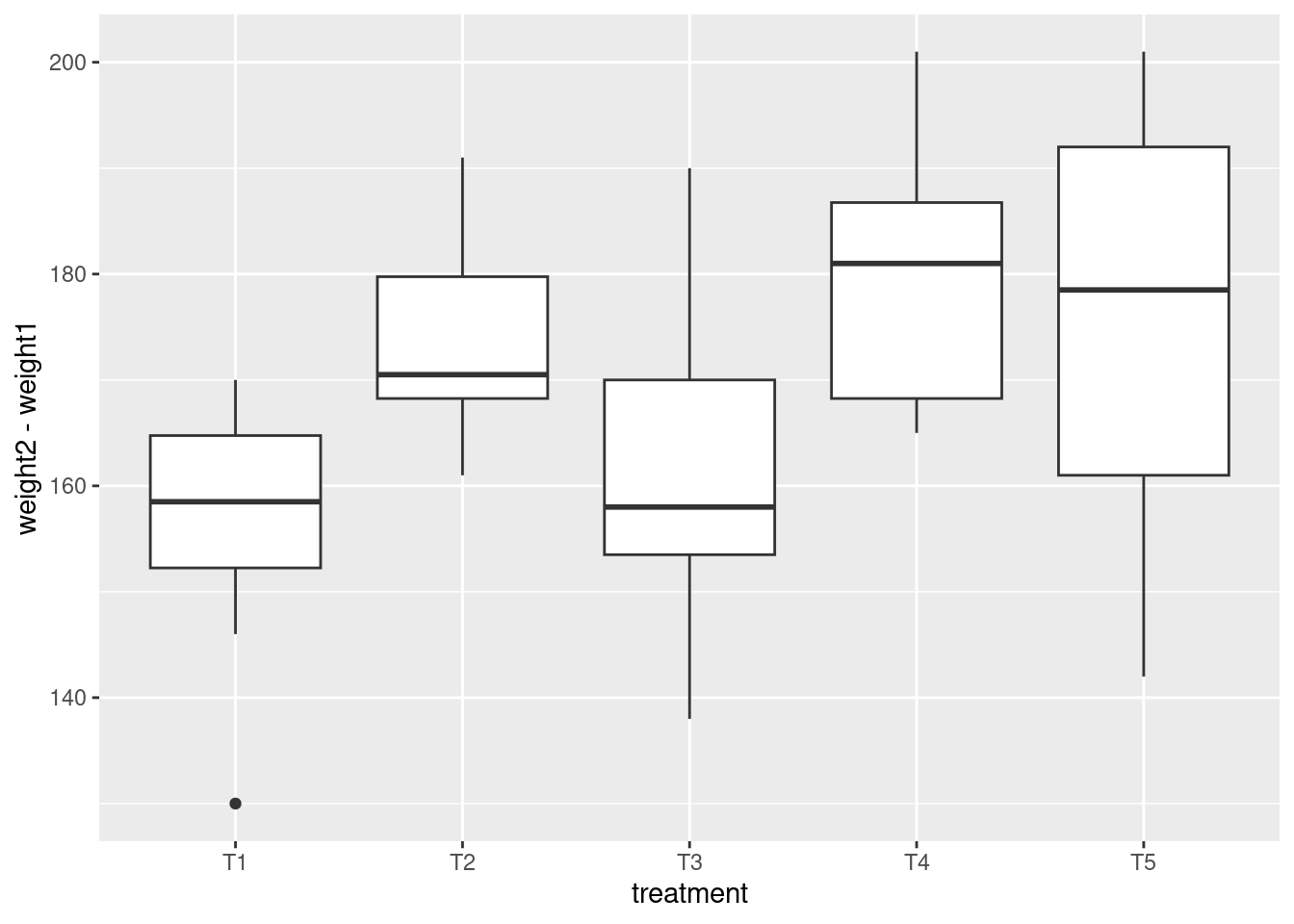
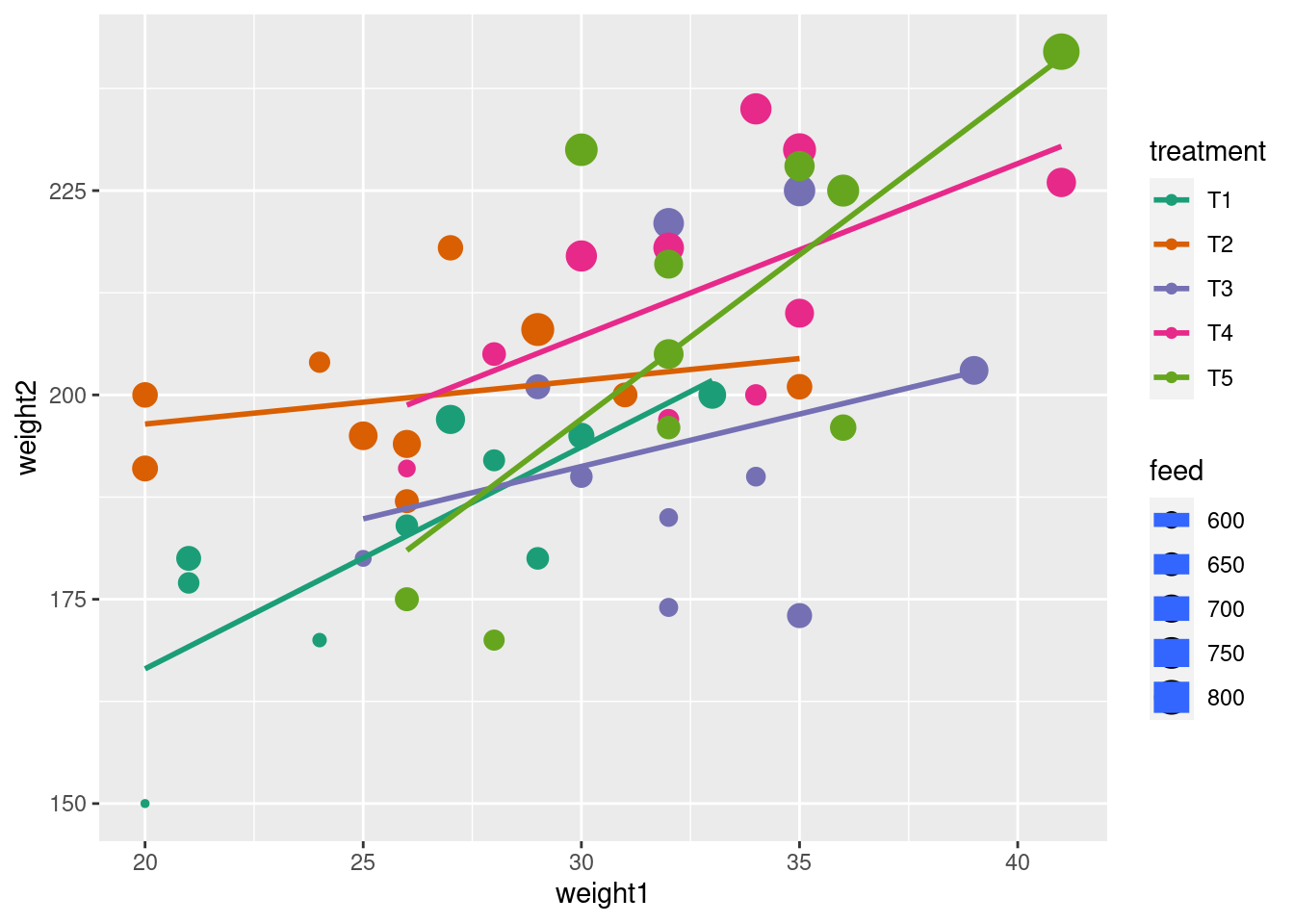
library(tidyverse) # includes `ggplot2`
data(crampton.pig, package = "agridat")
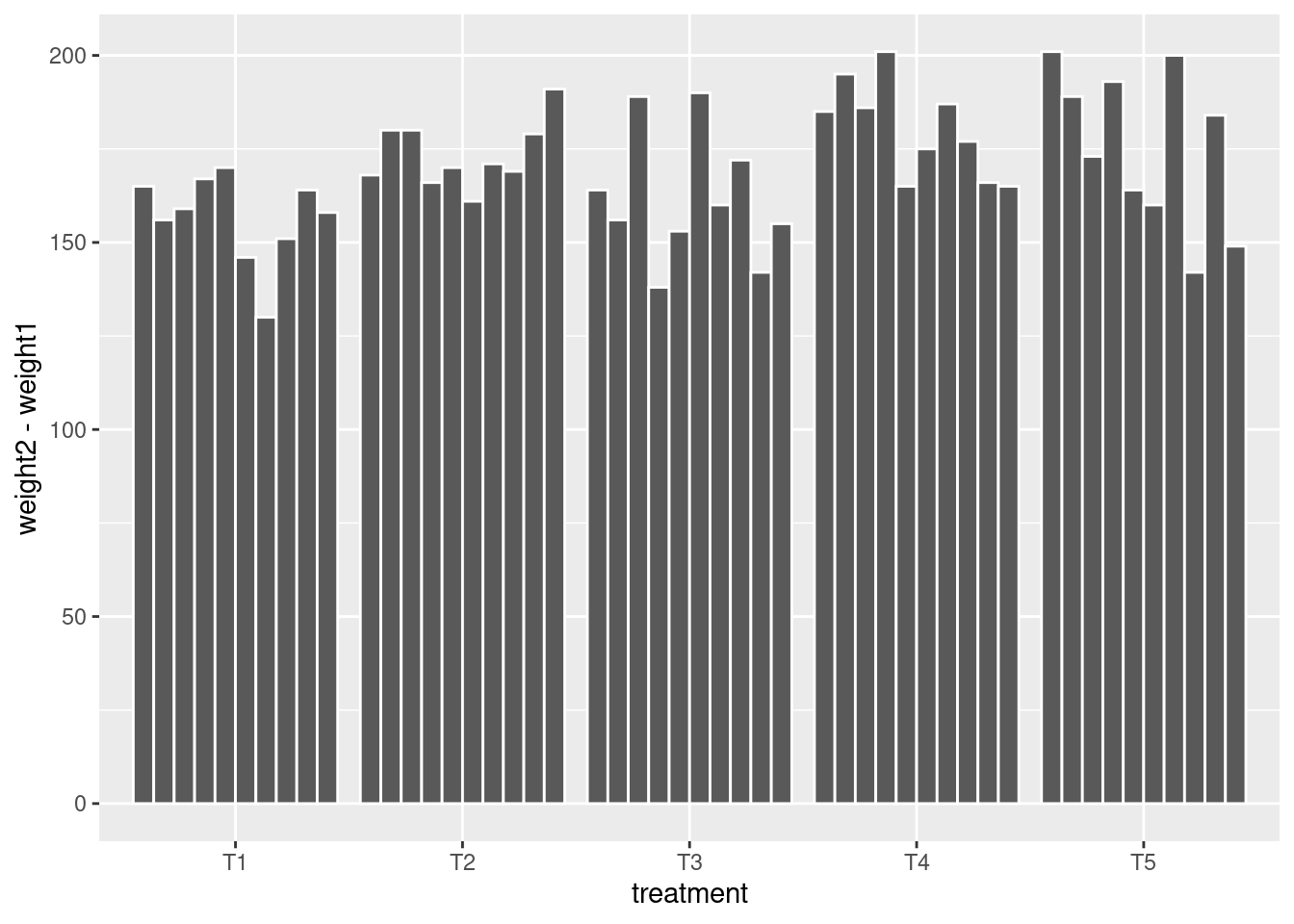
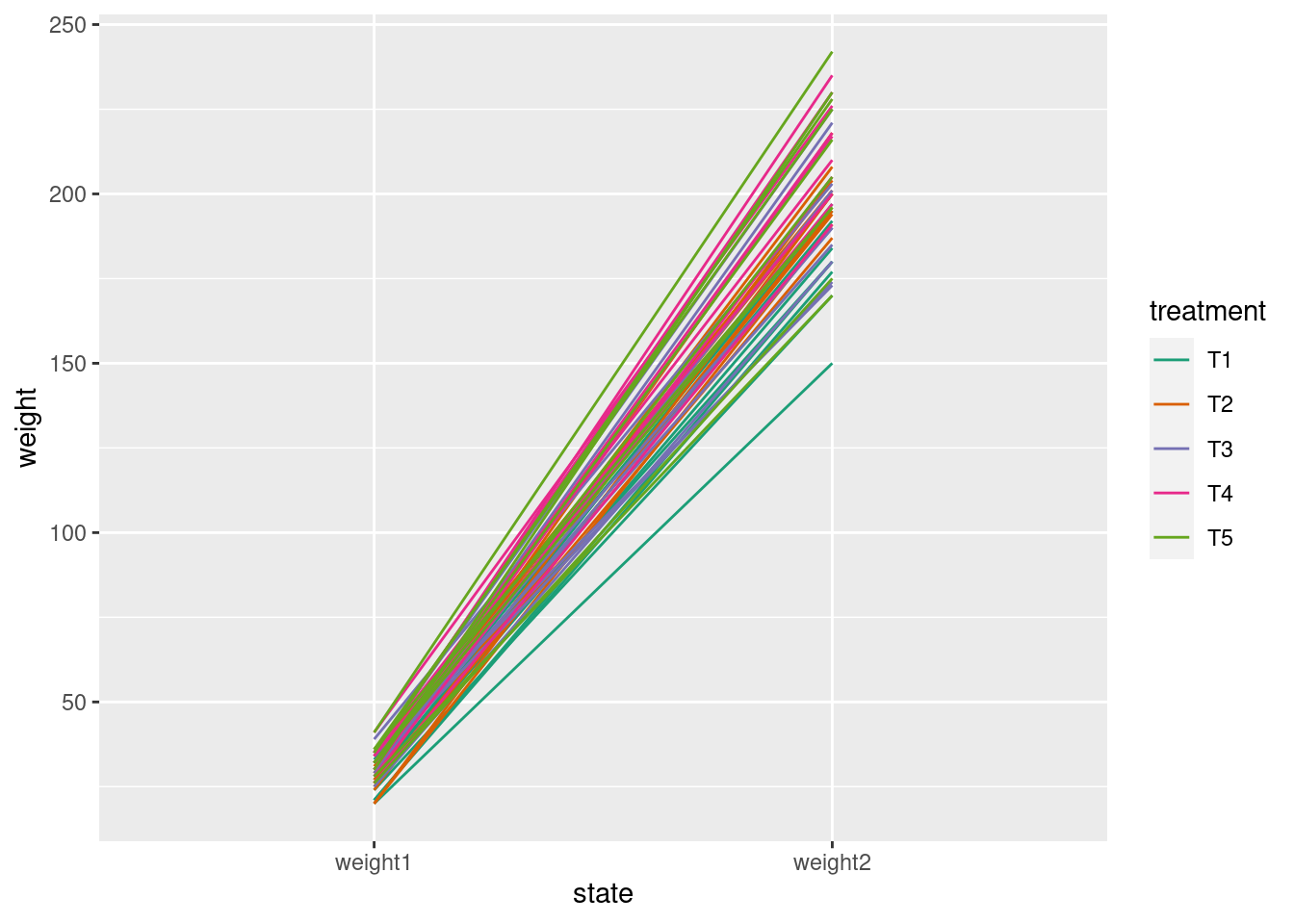
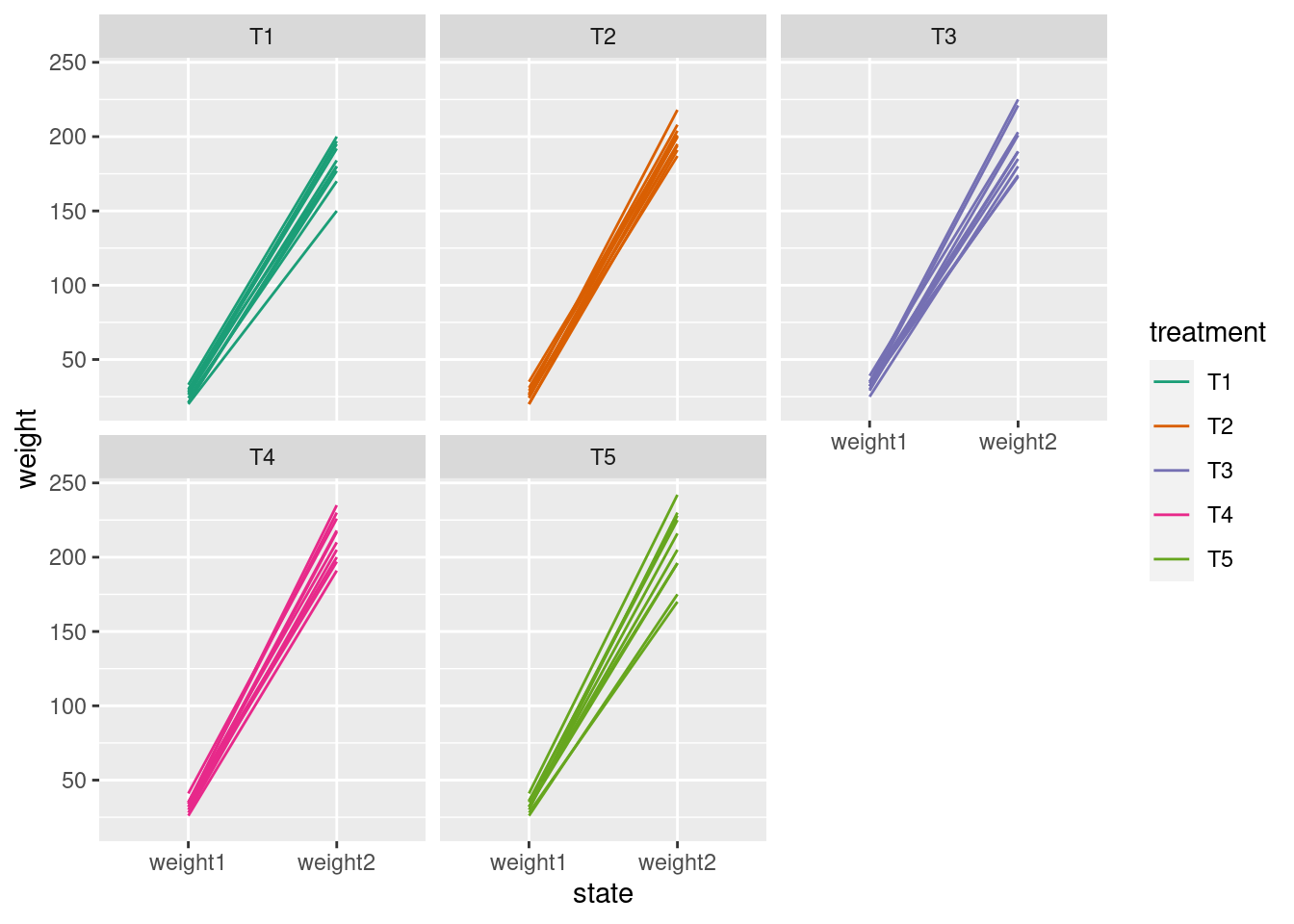
glimpse(crampton.pig)Rows: 50
Columns: 5
$ treatment <fct> T1, T1, T1, T1, T1, T1, T1, T1, T1, T1, T2, T2, T2, T2, T2, …
$ rep <fct> R1, R2, R3, R4, R5, R6, R7, R8, R9, R10, R1, R2, R3, R4, R5,…
$ weight1 <int> 30, 21, 21, 33, 27, 24, 20, 29, 28, 26, 26, 24, 20, 35, 25, …
$ feed <int> 674, 628, 661, 694, 713, 585, 575, 638, 632, 637, 699, 626, …
$ weight2 <int> 195, 177, 180, 200, 197, 170, 150, 180, 192, 184, 194, 204, …