class: split-60 title-slide2 with-border white background-image: url("images/bg1.jpg") background-size: cover .column.shade_main[.content[ <br><br> # Statistical Methods for Omics Assisted Breeding ## <b style='color:#FFEB3B'>Data Visualization in <span><i class="fab fa-r-project faa-none animated "></i></span></b> ### <br>Emi Tanaka <br>emi.tanaka@sydney.edu.au <br>School of Mathematics and Statisitcs ### 2018/11/12 <br> ### <span style='font-size:14pt; color:pink!important'>These slides may take a while to render properly. You can find the pdf <a href="https://www.dropbox.com/s/hulz2ua8kszehvr/day1-session02-datavis.pdf?dl=1" download="day1-session02-datavis.pdf">here</a>.</span> <br><br> ### <a rel='license' href='http://creativecommons.org/licenses/by-sa/4.0/'><img alt='Creative Commons License' style='border-width:0; width:30pt' src='images/cc.svg' /><img alt='Creative Commons License' style='border-width:0; width:20pt' src='images/by.svg' /><img alt='Creative Commons License' style='border-width:0; width:20pt' src='images/sa.svg' /></a><span style='font-size:10pt'> This work by <span xmlns:cc='http://creativecommons.org/ns#' property='cc:attributionName'>Emi Tanaka</span> is licensed under a <a rel='license' href='http://creativecommons.org/licenses/by-sa/4.0/'>Creative Commons Attribution-ShareAlike 4.0 International License</a>.</span> ]] .column[.content[ ]] <img src="images/USydLogo-white.svg" style="position:absolute; top:80%; left:40%;width:200px"> <img src="images/anim.gif" style="position:absolute; top:43%; left:37%;width:240px"> --- class: split-70 with-border .column[.content[ # Data Visualisation in `R` * `R` has many contributed packages that extend from the standard `base` installation. * Today we will learn about `ggplot2` and `desplot` R packages with a light touch on `plotly`. # Why Data Visualisation in `R`? * You can make .indigo[publication quality] graphs. * The graphs are easily .indigo[reproducible]. <span><i class="fas fa-exclamation-triangle faa-flash animated " style=" color:red;"></i></span> .red[`ggplot2` is quite stable now but `R` packages are contributed and can change in future iterations]. ]] .column.bg-blue[.content.center.vmiddle[ <img src="images/rlogo.svg" width="80%"/> ]] --- class: bg-main1 split-30 hide-slide-number .column.bg-main3[ ] .column.slide-in-right[ .sliderbox.bg-main2.vmiddle[ # .font_large[`ggplot`] ]] --- class: split-70 with-border .column[.content[ # `ggplot2` R package * `ggplot2` is a powerful data visualisation R package with a large community following that is built on the .indigo[layered grammar of graphics] by Wickham (2008). * One of the reason that makes it powerful is because of its ease in extensibility resulting in many extension packages. * `ggplot2` uses `qplot` or `ggplot` to make graphics * `qplot` is useful for making quick graphs (especially when data is not in a `data.frame`) but `ggplot` is advisable for most occasions. * We will only cover `ggplot`. * To get started, load the package: ```r library(ggplot2) # or library(tidyverse) ``` .bottom_abs.width100.font_small[ Wickham (2008) Practical tools for exploring data and models. PhD Thesis. ] ]] .column.bg-blue[.content.center.vmiddle[ <img src="images/ggplot2.png" width="80%"/> ]] --- class: split-70 with-border .column[.content[ # Layered Grammar of Graphics * Every `ggplot2` object has three key components: 1. .indigo[data], 1. A set of .indigo[aesthestic mapping] between variables in the data and visual properties (e.g color, size etc) 1. At least one .indigo[layer] describing how to render each observation; usually created with .indigo[geom] function. ```r str(iris) ``` ``` ## 'data.frame': 150 obs. of 5 variables: ## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ... ## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ... ## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ... ## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ... ## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ... ``` ]] .column.bg-blue[.content[ ```r ggplot(data=iris) + aes(x=Sepal.Length, y=Sepal.Width) + geom_point() ``` <img src="images/irisplot.svg" width="100%"> ]] --- class: split-70 with-border .column[.content[ # Every .black[layer] has: 1. `geom` - the geometric object to use display the data, and `stat` - statistical transformation to use on the data for this layer. 1. .indigo[data] and .indigo[mapping] (aesthestics) which is usually inherited from `ggplot()` object. 1. `position` - position in the coordinate system. ```r p <- ggplot(iris, aes(Sepal.Length, Sepal.Width)) p + geom_point() # blank + geom layer ``` which is a short-hand for: ```r p + layer(geom="point", stat="identity", position="identity") ``` ]] .column.bg-blue[.content[ Every `ggplot` object has: 1. Data 1. Aesthesitc mapping 1. Layer(s) Purpose of a layer is to display: * the raw .yellow[data], * a .yellow[statistical summary], or * additional .yellow[metadata] such as context, annotations, and references. ]] --- class: split-40 .row[ .split-50[ .column[.content[ # Some `geom` objects ```r p <- ggplot(iris, aes(Species, Sepal.Width)) class(p) ``` ``` ## [1] "gg" "ggplot" ``` ]] .column[.content.vmiddle[ .img-fill[] <br> .font_small[ Image source:<br> http://suruchifialoke.com/2016-10-13-machine-learning-tutorial-iris-classification/ ] ]]] ] .row[ .split-four[ .column.center[ ```r p + geom_blank() ``` <!-- --> ] .column.center[ ```r p + geom_point() ``` <!-- --> ] .column.center[ ```r p + geom_boxplot() ``` <!-- --> ] .column.center[ ```r p + geom_violin() ``` <!-- --> ] ]] --- class: split-20 .row[.content.nopadding[ ## Drawing lines ```r p <- ggplot(iris, aes(Petal.Length, Petal.Width)) + geom_point(colour="gray") ``` ]] .row[ .split-two[ .row[ .split-two[ .column.center[ ```r p + geom_abline(intercept=-0.4,slope=0.4) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-13-1.png" style="display: block; margin: auto;" /> ] .column.center[ ```r p + geom_smooth(method="lm") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-14-1.png" style="display: block; margin: auto;" /> ] ]] .row[ .split-two[ .column.center[ ```r p + geom_hline(yintercept=0) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-15-1.png" style="display: block; margin: auto;" /> ] .column.center[ ```r p + geom_vline(xintercept=0) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-16-1.png" style="display: block; margin: auto;" /> ] ]]]] --- class: split-20 .row[.content.nopadding[ # Distribution by group ```r p <- ggplot(iris, aes(Petal.Width, fill=Species)) ``` ]] .row[ .split-two[ .row[ .split-two[ .column.center[ ```r p + geom_dotplot() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-18-1.png" style="display: block; margin: auto;" /> ] .column.center[ ```r p + geom_histogram() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-19-1.png" style="display: block; margin: auto;" /> ] ]] .row[ .split-two[ .column.center[ ```r p + geom_density() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-20-1.png" style="display: block; margin: auto;" /> ] .column.center[ ```r p + geom_freqpoly(aes(color=Species)) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-21-1.png" style="display: block; margin: auto;" /> ] ]]]] --- class: split-70 with-border .column[.content.pad10px[ .font_sm55[ <table class="table table-striped table-hover table-condensed table-responsive" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> geom </th> <th style="text-align:left;"> Description </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> geom_abline </td> <td style="text-align:left;"> Reference lines: horizontal, vertical, and diagonal </td> </tr> <tr> <td style="text-align:left;"> geom_bar </td> <td style="text-align:left;"> Bar charts </td> </tr> <tr> <td style="text-align:left;"> geom_bin2d </td> <td style="text-align:left;"> Heatmap of 2d bin counts </td> </tr> <tr> <td style="text-align:left;"> geom_blank </td> <td style="text-align:left;"> Draw nothing </td> </tr> <tr> <td style="text-align:left;"> geom_boxplot </td> <td style="text-align:left;"> A box and whiskers plot (in the style of Tukey) </td> </tr> <tr> <td style="text-align:left;"> geom_contour </td> <td style="text-align:left;"> 2d contours of a 3d surface </td> </tr> <tr> <td style="text-align:left;"> geom_count </td> <td style="text-align:left;"> Count overlapping points </td> </tr> <tr> <td style="text-align:left;"> geom_density </td> <td style="text-align:left;"> Smoothed density estimates </td> </tr> <tr> <td style="text-align:left;"> geom_density_2d </td> <td style="text-align:left;"> Contours of a 2d density estimate </td> </tr> <tr> <td style="text-align:left;"> geom_dotplot </td> <td style="text-align:left;"> Dot plot </td> </tr> <tr> <td style="text-align:left;"> geom_errorbarh </td> <td style="text-align:left;"> Horizontal error bars </td> </tr> <tr> <td style="text-align:left;"> geom_hex </td> <td style="text-align:left;"> Hexagonal heatmap of 2d bin counts </td> </tr> <tr> <td style="text-align:left;"> geom_freqpoly </td> <td style="text-align:left;"> Histograms and frequency polygons </td> </tr> <tr> <td style="text-align:left;"> geom_jitter </td> <td style="text-align:left;"> Jittered points </td> </tr> <tr> <td style="text-align:left;"> geom_crossbar </td> <td style="text-align:left;"> Vertical intervals: lines, crossbars & errorbars </td> </tr> <tr> <td style="text-align:left;"> geom_map </td> <td style="text-align:left;"> Polygons from a reference map </td> </tr> <tr> <td style="text-align:left;"> geom_path </td> <td style="text-align:left;"> Connect observations </td> </tr> <tr> <td style="text-align:left;"> geom_point </td> <td style="text-align:left;"> Points </td> </tr> <tr> <td style="text-align:left;"> geom_polygon </td> <td style="text-align:left;"> Polygons </td> </tr> <tr> <td style="text-align:left;"> geom_qq_line </td> <td style="text-align:left;"> A quantile-quantile plot </td> </tr> <tr> <td style="text-align:left;"> geom_quantile </td> <td style="text-align:left;"> Quantile regression </td> </tr> <tr> <td style="text-align:left;"> geom_ribbon </td> <td style="text-align:left;"> Ribbons and area plots </td> </tr> <tr> <td style="text-align:left;"> geom_rug </td> <td style="text-align:left;"> Rug plots in the margins </td> </tr> <tr> <td style="text-align:left;"> geom_segment </td> <td style="text-align:left;"> Line segments and curves </td> </tr> <tr> <td style="text-align:left;"> geom_smooth </td> <td style="text-align:left;"> Smoothed conditional means </td> </tr> <tr> <td style="text-align:left;"> geom_spoke </td> <td style="text-align:left;"> Line segments parameterised by location, direction and distance </td> </tr> <tr> <td style="text-align:left;"> geom_label </td> <td style="text-align:left;"> Text </td> </tr> <tr> <td style="text-align:left;"> geom_raster </td> <td style="text-align:left;"> Rectangles </td> </tr> <tr> <td style="text-align:left;"> geom_violin </td> <td style="text-align:left;"> Violin plot </td> </tr> </tbody> </table> ]]] .column.bg-blue[.content.vmiddle.white.center[ # .font-mono.font_large[geom] ]] --- class: split-70 with-border .column[.content.font_sm80[ # Statistical Tranformation ```r head(iris[, c("Petal.Width", "Species")]) # raw data ``` ``` ## Petal.Width Species ## 1 0.2 setosa ## 2 0.2 setosa ## 3 0.2 setosa ## 4 0.2 setosa ## 5 0.2 setosa ## 6 0.4 setosa ``` `stat_bin(bins=7, mapping=aes(Petal.Width, fill=Species))` <span><i class="fas fa-arrow-down faa-none animated "></i></span> Under the hood, the raw data is transformed into statistics and this is passed onto the `geom` where here `geom="bar"` is default. ``` ## fill y count x xmin xmax density ncount ## 1 #619CFF 0 0 0.0 -0.2 0.2 0.0 0.0000000 ## 2 #00BA38 0 0 0.0 -0.2 0.2 0.0 0.0000000 ## 3 #F8766D 34 34 0.0 -0.2 0.2 1.7 1.0000000 ## 4 #619CFF 0 0 0.4 0.2 0.6 0.0 0.0000000 ## 5 #00BA38 0 0 0.4 0.2 0.6 0.0 0.0000000 ## 6 #F8766D 16 16 0.4 0.2 0.6 0.8 0.4705882 ``` ]] .column.bg-yellow[.vmiddle[ <!-- --> ]] --- class: split-20 .row[.content[ # Using `stat` with different `geom` object ```r p <- ggplot(iris, aes(Petal.Width, fill=Species)) ``` ]] .row[ .split-two[ .row[ .split-two[ .column.center[ ```r p + stat_bin() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-27-1.png" style="display: block; margin: auto;" /> ] .column.center[ ```r p + stat_bin(geom="bar") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-28-1.png" style="display: block; margin: auto;" /> ] ]] .row[ .split-two[ .column.center[ ```r p + stat_bin(geom="point") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-29-1.png" style="display: block; margin: auto;" /> ] .column.center[ ```r p + stat_bin(geom="line") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-30-1.png" style="display: block; margin: auto;" /> ] ]]]] --- class: split-70 with-border .column[.content.pad10px[ .font_sm74[ <table class="table table-striped table-hover table-condensed table-responsive" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> stat </th> <th style="text-align:left;"> Description </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> stat_count </td> <td style="text-align:left;"> Bar charts </td> </tr> <tr> <td style="text-align:left;"> stat_bin_2d </td> <td style="text-align:left;"> Heatmap of 2d bin counts </td> </tr> <tr> <td style="text-align:left;"> stat_boxplot </td> <td style="text-align:left;"> A box and whiskers plot (in the style of Tukey) </td> </tr> <tr> <td style="text-align:left;"> stat_contour </td> <td style="text-align:left;"> 2d contours of a 3d surface </td> </tr> <tr> <td style="text-align:left;"> stat_sum </td> <td style="text-align:left;"> Count overlapping points </td> </tr> <tr> <td style="text-align:left;"> stat_density </td> <td style="text-align:left;"> Smoothed density estimates </td> </tr> <tr> <td style="text-align:left;"> stat_density_2d </td> <td style="text-align:left;"> Contours of a 2d density estimate </td> </tr> <tr> <td style="text-align:left;"> stat_bin_hex </td> <td style="text-align:left;"> Hexagonal heatmap of 2d bin counts </td> </tr> <tr> <td style="text-align:left;"> stat_bin </td> <td style="text-align:left;"> Histograms and frequency polygons </td> </tr> <tr> <td style="text-align:left;"> stat_qq_line </td> <td style="text-align:left;"> A quantile-quantile plot </td> </tr> <tr> <td style="text-align:left;"> stat_quantile </td> <td style="text-align:left;"> Quantile regression </td> </tr> <tr> <td style="text-align:left;"> stat_smooth </td> <td style="text-align:left;"> Smoothed conditional means </td> </tr> <tr> <td style="text-align:left;"> stat_spoke </td> <td style="text-align:left;"> Line segments parameterised by location, direction and distance </td> </tr> <tr> <td style="text-align:left;"> stat_ydensity </td> <td style="text-align:left;"> Violin plot </td> </tr> <tr> <td style="text-align:left;"> stat_sf </td> <td style="text-align:left;"> Visualise sf objects </td> </tr> <tr> <td style="text-align:left;"> stat_ecdf </td> <td style="text-align:left;"> Compute empirical cumulative distribution </td> </tr> <tr> <td style="text-align:left;"> stat_ellipse </td> <td style="text-align:left;"> Compute normal confidence ellipses </td> </tr> <tr> <td style="text-align:left;"> stat_function </td> <td style="text-align:left;"> Compute function for each x value </td> </tr> <tr> <td style="text-align:left;"> stat_identity </td> <td style="text-align:left;"> Leave data as is </td> </tr> <tr> <td style="text-align:left;"> stat_sf_coordinates </td> <td style="text-align:left;"> Extract coordinates from 'sf' objects </td> </tr> <tr> <td style="text-align:left;"> stat_summary_bin </td> <td style="text-align:left;"> Summarise y values at unique/binned x </td> </tr> <tr> <td style="text-align:left;"> stat_summary_2d </td> <td style="text-align:left;"> Bin and summarise in 2d (rectangle & hexagons) </td> </tr> <tr> <td style="text-align:left;"> stat_unique </td> <td style="text-align:left;"> Remove duplicates </td> </tr> </tbody> </table> ]]] .column.bg-blue[.content.vmiddle.white.center[ # .font-mono.font_large[stat] ]] --- class: bg-main1 split-30 hide-slide-number .column.bg-main3[ ] .column.slide-in-right[ .sliderbox.bg-main2.vmiddle[ # .font_large[Customisation] ]] --- class: split-two with-border .column[.content[ # Changing Color There are many color palettes available, e.g. ```r library(RColorBrewer) ggplot(iris, aes(Petal.Width, * fill=Species)) + geom_dotplot() + * scale_fill_brewer(palette="Set3") ``` <!-- --> ]] .column[ <!-- --> ] --- class: split-two with-border .column[.content[ # Grey-scale ```r ggplot(iris, aes(Petal.Width, fill=Species)) + geom_dotplot() + * scale_fill_grey() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-34-1.png" style="display: block; margin: auto;" /> ]] .column[.content[ # Manual scale .font_sm85[ ```r ggplot(iris, aes(Petal.Width, fill=Species)) + geom_dotplot() + *scale_fill_manual( * values=c("red","blue", "green"), * labels=c("setosa", "versicolor", "virginica")) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-35-1.png" style="display: block; margin: auto;" /> ]]] --- class: split-two with-border .column[.content[ # Color variable is `factor` .font_sm60[ ```r ggplot(iris, aes(Petal.Width, Petal.Length, * color=Species)) + geom_point(size=2) + * scale_color_brewer(palette="Set1") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-36-1.png" style="display: block; margin: auto;" /> ]]] .column[.content[ # Color variable is continuous .font_sm60[ ```r ggplot(iris, aes(Petal.Width, Petal.Length, * color=Sepal.Length)) + geom_point(size=2) + * scale_color_distiller(palette="YlGnBu") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-37-1.png" style="display: block; margin: auto;" /> ]]] --- class: split-70 with-border .column[.content[ ## Counts of yellow/white and sweet/starchy maize kernels .font_sm60[ Here I am massaging the data to get the counts for the 8th maize ear by observer 1 (plant pathologist): ```r library(agridat); library(dplyr) (maize <- pearl.kernels %>% filter(ear=="Ear08" & obs=="Obs01") %>% select(ys, yt, ws, wt) %>% tidyr::gather("Type", "Count", ys:wt) %>% mutate(Color=case_when( Type %in% c("ys", "yt") ~ "Yellow", Type %in% c("ws", "wt") ~ "White" ),Kernel=case_when( Type %in% c("ys", "ws") ~ "Starchy", Type %in% c("yt", "wt") ~ "Sweet"))) ``` ``` ## Type Count Color Kernel ## 1 ys 352 Yellow Starchy ## 2 yt 102 Yellow Sweet ## 3 ws 52 White Starchy ## 4 wt 26 White Sweet ``` ] .bottom_abs.width100.font_small[ Pearl, Raymond (1911) The Personal Equation In Breeding Experiments Involving Certain Characters of Maize *Biological Bulletin* **21** 339-366 ] ]] .column.bg-blue[.content.vmiddle.nopadding[ .img-fill[] .font_small[Image source: http://corncommentary.com/2012/05/22/using-the-kfc-kernel-for-cellulosic/ ]]] --- class: split-70 with-border .column[.content[ # Example: Observer 1 for Maize Ear 8 ```r p <- ggplot(maize, aes(Kernel, Count, fill=Color)) + geom_bar(stat="identity") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-40-1.png" style="display: block; margin: auto;" /> ]] .column.bg-blue[.content.vmiddle[ <img src="images/corn-farm.jpg" width="114%" style="margin-right: -7%; margin-left: -7%;"> .font_small[ Image Source:<br> https://agrifarmingtips.com/maize-cultivation-process/] ]] --- class: split-70 with-border count: false .column[.content[ # Example: Observer 1 for Maize Ear 8 ```r p <- ggplot(maize, aes(Kernel, Count, fill=Color)) + geom_bar(stat="identity", color="black") + scale_fill_manual(values=c("white", "yellow"), label=c("White", "Yellow")) + guides(fill=FALSE) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-42-1.png" style="display: block; margin: auto;" /> ]] .column.bg-blue[.content.vmiddle[ <img src="images/corn-farm.jpg" width="114%" style="margin-right: -7%; margin-left: -7%;"> .font_small[ Image Source:<br> https://agrifarmingtips.com/maize-cultivation-process/] ]] --- class: split-70 with-border count: false .column[.content[ # Example: Observer 1 for Maize Ear 8 <pre> <code class="r hljs remark-code"><div class="remark-code-line">p <- ggplot(maize, aes(Kernel, Count, fill=Color)) + </div> <div class="remark-code-line"><s> geom_bar(stat=<span class="hljs-string">"identity"</span>, color=<span class="hljs-string">"black"</span>) + </s></div> <div class="remark-code-line"> scale_fill_manual(values=c(<span class="hljs-string">"white"</span>, <span class="hljs-string">"yellow"</span>), </div> <div class="remark-code-line"> label=c(<span class="hljs-string">"White"</span>, <span class="hljs-string">"Yellow"</span>)) + </div> <div class="remark-code-line"> guides(fill=<span class="hljs-literal">FALSE</span>)</div> </code></pre> <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-43-1.png" style="display: block; margin: auto;" /> ]] .column.bg-blue[.content.vmiddle[ <img src="images/corn-farm.jpg" width="114%" style="margin-right: -7%; margin-left: -7%;"> .font_small[ Image Source:<br> https://agrifarmingtips.com/maize-cultivation-process/] ]] --- class: split-two .row[ .split-two[ .column[.content[ ## Position for `geom_bar` .center[ ```r p + geom_bar() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-45-1.png" style="display: block; margin: auto;" /> ]]] .column[.content.center[ ## ```r p + geom_bar(position="stack") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-47-1.png" style="display: block; margin: auto;" /> ]]]] .row[ .split-two[ .column[.content.center[ ```r p + geom_bar(position="dodge") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-49-1.png" style="display: block; margin: auto;" /> ]] .column[.content.center[ ```r p + geom_bar(position="fill") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-51-1.png" style="display: block; margin: auto;" /> ]]]] .bottom_abs.width100.font_sm[All `geom_bar` include the arguments `stat="identity"` and `color="black"`.] --- class: split-20 .row[.content[ ## Coordinate system ```r p2 <- ggplot(maize, aes(1, Count, fill=Type)) + guides(fill=FALSE) + theme_void() ``` ]] .row[.content[ .split-two[ .row[ .split-two[ .column[.content[ .center[ ```r p + geom_bar() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-54-1.png" style="display: block; margin: auto;" /> ]]] .column[.content.center[ ```r p + geom_bar() + coord_polar(theta="y") ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-56-1.png" style="display: block; margin: auto;" /> ]]]] .row[ .split-two[ .column[.content.center[ ```r p + geom_bar() + coord_flip() ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-58-1.png" style="display: block; margin: auto;" /> ]] .column[.content.center[ ```r p + geom_bar() + coord_polar(theta="y", direction=-1) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-60-1.png" style="display: block; margin: auto;" /> ]]]]]]] .bottom_abs.width100.font_small[All `geom_bar` include the arguments `stat="identity"` and `color="black"`.] --- class: split-20 .row[.content[ ## Overplotting ```r g <- ggplot(pearl.kernels, aes(ear, ys, color=ear, size=3)) + xlab(NULL) + guides(color=FALSE, size=FALSE) + ylab("No. of Yellow\n Starchy Kernel") ``` ]] .row[ .split-two[ .row[ .split-two[ .column[.content.center[ ```r g + geom_point() ``` <!-- --> ]] .column[.content.center[ ```r g + geom_point(position="jitter") ``` <!-- --> ]]]] .row[ .split-two[ .column[.content.center[ ```r g + geom_point(alpha=1 / 3) ``` <!-- --> ]] .column[.content.center[ ```r g + geom_point(alpha=1 / 6) ``` <!-- --> ]]]] ]] --- class: split-two .row[.split-70[ .column[.content[ # Massaging data to tidy form ```r maize2 <- pearl.kernels %>% tidyr::gather("Type", "Count", ys:wt) %>% mutate(Color=ifelse(substr(Type, 1, 1)=="y", "Yellow", "White"), Kernel=ifelse(substr(Type, 2, 2)=="s", "Starchy", "Sweet"), obs=factor(as.integer(substring(obs, 4, 5)))) ``` ]] .column[.content[ .img-fill[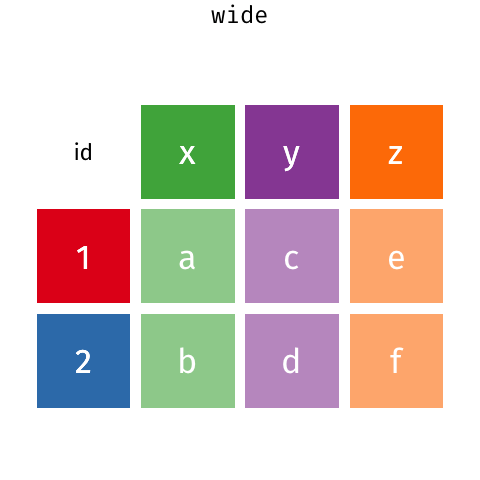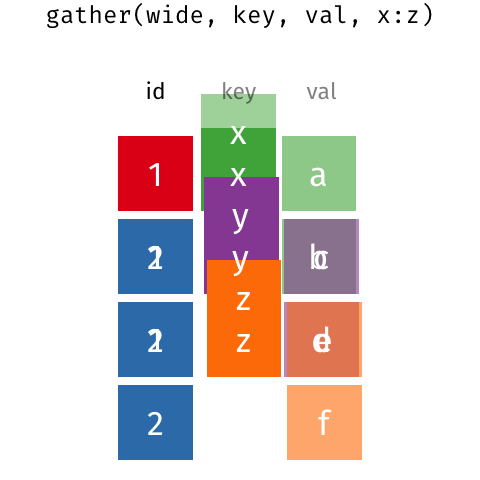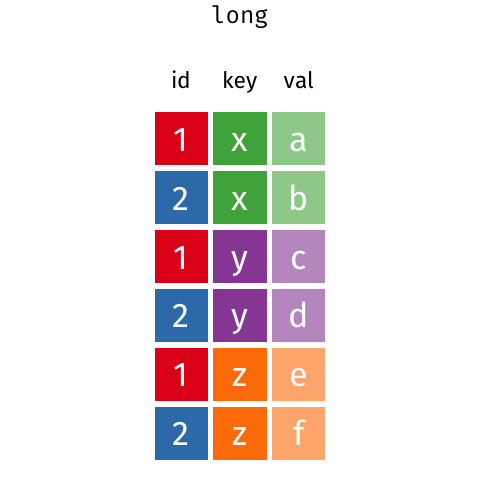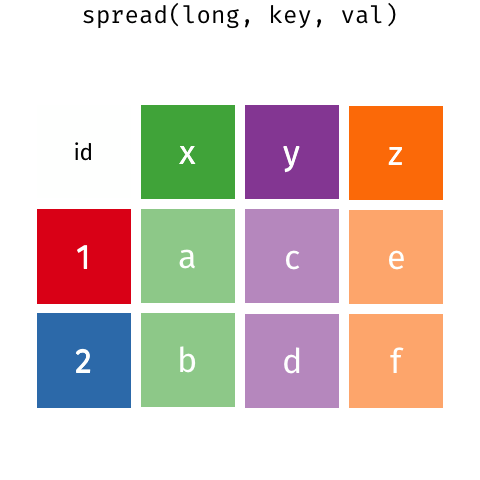] ]] ]] .row[.split-two[ .column[.content[ ```r head(maize2) ``` ``` ear obs Type Count Color Kernel 1 Ear08 1 ys 352 Yellow Starchy 2 Ear08 2 ys 322 Yellow Starchy 3 Ear08 3 ys 298 Yellow Starchy 4 Ear08 4 ys 332 Yellow Starchy 5 Ear08 5 ys 305 Yellow Starchy 6 Ear08 6 ys 313 Yellow Starchy ``` ]] .column[.content[ ```r head(pearl.kernels) ``` ``` ear obs ys yt ws wt 1 Ear08 Obs01 352 102 52 26 2 Ear08 Obs02 322 49 82 79 3 Ear08 Obs03 298 75 108 51 4 Ear08 Obs04 332 101 71 28 5 Ear08 Obs05 305 101 86 40 6 Ear08 Obs06 313 100 90 29 ``` ]] ]] --- class: split-two .column[.content[ # Faceting ```r ggplot(maize2, aes(obs, Count, fill=Type)) + geom_bar(stat="identity") + xlab("Observer") + * facet_wrap(~ear) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-69-1.png" style="display: block; margin: auto;" /> ]] .column[.content[ # ```r ear8 <- maize2 %>% filter(ear=="Ear08") %>% ggplot(aes(obs, Count, fill=Type)) + geom_bar(stat="identity", show.legend=F) + labs(tag="(A)", title="Ear 8", x="Observer") + * facet_grid(Color ~ Kernel) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-71-1.png" style="display: block; margin: auto;" /> ]] --- class: split-70 .column[.content[ # Patching Plots Together ```r library(patchwork) ear8 + ear9 + ear10 + ear11 + plot_layout(ncol = 2) ``` <!-- --> ]] .column.bg-blue[.content.vmiddle.center[ <img src="images/patchwork.png" width="80%"> ]] --- class: split-50 with-border .column[.content[ # Example: Barley in Norway .font_sm80[Average height each year for 15 genotypes of barley in Norway from 1974-1982.] ```r str(aastveit.barley.height) # from agridat ``` ``` ## 'data.frame': 135 obs. of 3 variables: ## $ year : int 1974 1975 1976 1977 1978 1979 1980 1981 1982 1974 ... ## $ gen : Factor w/ 15 levels "G01","G02","G03",..: 1 1 1 1 1 1 1 1 1 2 ... ## $ height: num 81 67.3 71.5 64.3 55.8 84.9 86.2 88 72 72.3 ... ``` .font_sm80[The covariate information are found in `aastveit.barley.covs`.<br> Use below command to find information about the data: ```r ?aastveit.barley.height ?aastveit.barley.covs ``` ] .bottom_abs.width100.font_small[ Aastveit, A. H. and Martens, H. (1986). ANOVA interactions interpreted by partial least squares regression. *Biometrics* **42** 829–844. ]]] .column.bg-white[.content[ ```r barley <- aastveit.barley.height %>% left_join(aastveit.barley.covs, by="year") ``` <img src="images/left-join.gif"> ]] --- class: split-50 with-border .column[.content[ # Subset data for labels ```r (maxh_df <- barley %>% select(year, height, gen, T4) %>% group_by(year) %>% filter(height==max(height)) %>% arrange(year)) ``` ``` ## # A tibble: 9 x 4 ## # Groups: year [9] ## year height gen T4 ## <int> <dbl> <fct> <dbl> ## 1 1974 97 G15 12.1 ## 2 1975 83.3 G15 16.0 ## 3 1976 86.8 G15 17.4 ## 4 1977 80 G10 17.4 ## 5 1978 75.5 G11 13.9 ## 6 1979 97 G14 14.0 ## 7 1980 106. G11 13.9 ## 8 1981 106. G07 13.6 ## 9 1982 90.3 G14 11.6 ``` ]] .column.bg-white[.content[ # Labels with `geom_label` ```r g <- ggplot(barley, aes(T4, height)) + geom_point(size=4, aes(color=factor(year))) + guides(color=FALSE) + xlab("Avg temp (Celsius) in the 4-th period") + ylab("Height") g + geom_label(data=maxh_df, size=4, aes(T4, height, label=year)) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-78-1.png" style="display: block; margin: auto;" /> ]] --- class: split-two with-border .column[.content[ # Nudge labels + `geom_text` ```r g + * geom_text( data=maxh_df, size=4, * nudge_y=10, aes(T4, height, label=year)) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-79-1.png" style="display: block; margin: auto;" /> ]] .column[.content[ # `ggrepel` ```r library(ggrepel) g + * geom_label_repel( data=maxh_df, size=4, aes(T4, height, label=year)) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-80-1.png" style="display: block; margin: auto;" /> ]] --- class: split-50 with-border .column[.content[ # Annotation Text ```r g + annotate("text", x=12, y=100, label="1974", size=12) ``` <!-- --> ]] .column[.content[ # Annotation Rectangle ```r g + annotate("rect", xmin=15, xmax=18, ymin=-Inf, ymax=Inf, alpha=0.2, fill="red") ``` <!-- --> ]] --- class: split-60 with-border .column.bg-blue[.content[ # Changing Labels ```r g <- ggplot(vargas.wheat1.traits, aes(NGS, yield)) + geom_point(size=3) + geom_point(aes(colour=gen)) + geom_smooth(se=F, method="lm") + facet_wrap(~year) + * labs(colour="Genotype") + # changes the label name for color legend * labs(x="Number of grains per spikelet") + # same as xlab(..) * labs(y="Yield (kg/ha)") + # same as ylab(..) * labs(title="Durum Wheat at Ciudad Obregon, Mexico 1990-1995") + # same as ggtitle(..) * labs(subtitle="Source: Vargas et al. (1998) Interpreting Genotype x Environment Interaction in Wheat by Partial Least Squares Regression.") # same as ggtitle(subtitle=..) ``` ]] .column[.content.vmiddle.center[ <!-- --> ]] --- class: split-60 with-border .column.bg-blue[.content[ # Theme - customise the look ```r *g ``` ]] .column[.content.vmiddle.center[ <!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + *theme(legend.position="bottom") ``` ]] .column[.content.vmiddle.center[ <!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", *plot.title=element_text(face="bold", size=15)) ``` ]] .column[.content.vmiddle.center[ <!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), *plot.subtitle=element_text(face="italic", size=8)) ``` ]] .column[.content.vmiddle.center[ <!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), plot.subtitle=element_text(face="italic", size=8), *panel.background=element_rect(fill="white")) ``` ]] .column[.content.vmiddle.center[ 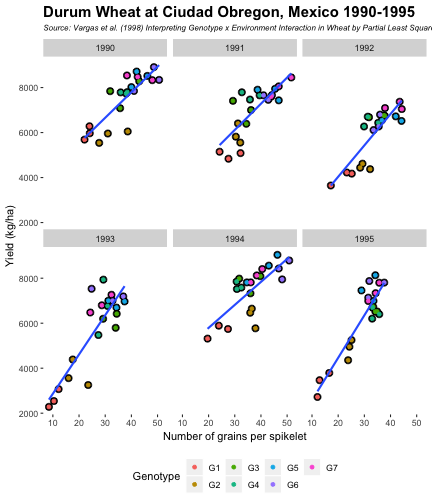<!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), plot.subtitle=element_text(face="italic", size=8), panel.background=element_rect(fill="white"), *panel.border=element_rect(colour="grey20", fill=NA)) ``` ]] .column[.content.vmiddle.center[ 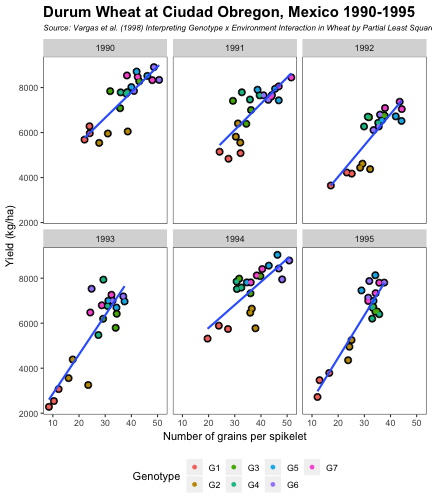<!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), plot.subtitle=element_text(face="italic", size=8), panel.background=element_rect(fill="white"), panel.border=element_rect(colour="grey20", fill=NA), *panel.grid=element_line(colour="grey92")) ``` ]] .column[.content.vmiddle.center[ 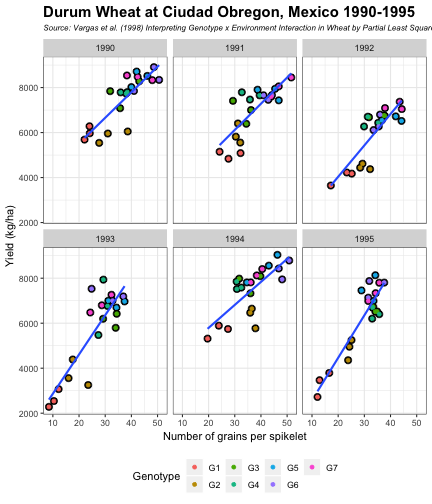<!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), plot.subtitle=element_text(face="italic", size=8), panel.background=element_rect(fill="white"), panel.border=element_rect(colour="grey20", fill=NA), panel.grid=element_line(colour="grey92"), *panel.grid.minor=element_line(size=rel(0.5))) ``` ]] .column[.content.vmiddle.center[ 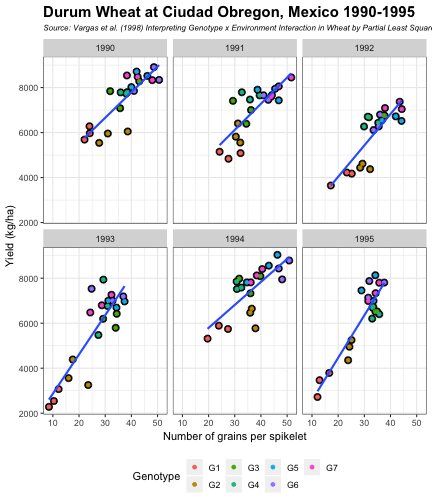<!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), plot.subtitle=element_text(face="italic", size=8), panel.background=element_rect(fill="white"), panel.border=element_rect(colour="grey20", fill=NA), panel.grid=element_line(colour="grey92"), panel.grid.minor=element_line(size=rel(0.5)), *strip.background=element_rect(fill="grey85", colour="grey20")) ``` ]] .column[.content.vmiddle.center[ 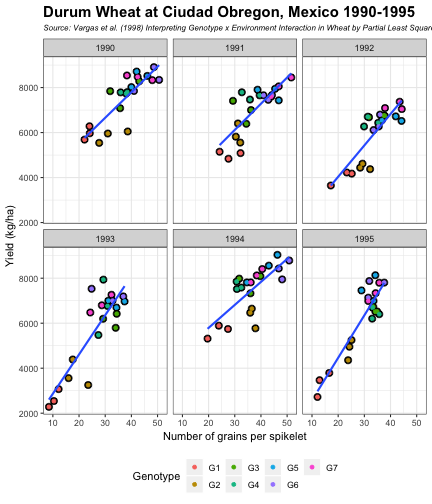<!-- --> ]] --- class: split-60 with-border count: false .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), plot.subtitle=element_text(face="italic", size=8), panel.background=element_rect(fill="white"), panel.border=element_rect(colour="grey20", fill=NA), panel.grid=element_line(colour="grey92"), panel.grid.minor=element_line(size=rel(0.5)), strip.background=element_rect(fill="grey85", colour="grey20"), *legend.key=element_rect(fill="white")) ``` ]] .column[.content.vmiddle.center[ 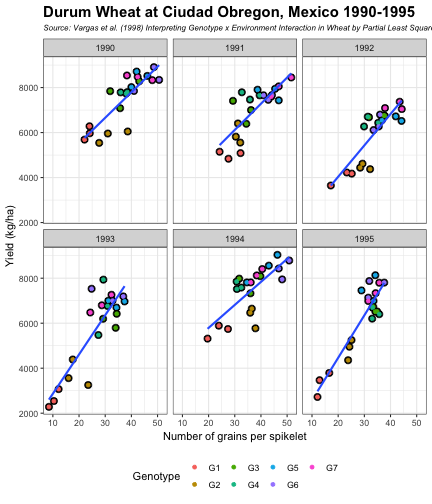<!-- --> ]] --- class: split-60 with-border .column.bg-blue[.content[ # Theme - customise the look ```r g + theme(legend.position="bottom", plot.title=element_text(face="bold", size=15), plot.subtitle=element_text(face="italic", size=8), panel.background=element_rect(fill="white"), panel.border=element_rect(colour="grey20", fill=NA), panel.grid=element_line(colour="grey92"), panel.grid.minor=element_line(size=rel(0.5)), strip.background=element_rect(fill="grey85", colour="grey20"), legend.key=element_rect(fill="white")) ``` or use a pre-defined theme: ```r g + *theme_bw() + theme(legend.position="bottom", plot.title=element_text(face="bold", size=14), plot.subtitle=element_text(face="italic", size=8)) ``` ]] .column[.content.vmiddle.center[ 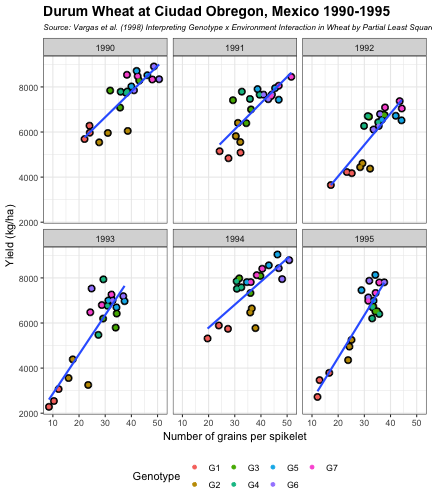<!-- --> ]] --- class: split-50 .column.bg-white[.content.center[ # More Pre-Defined Themes ```r g + theme_gray() ``` <!-- --> ```r g + theme_classic() ``` <!-- --> ]] .column.bg-white[.content.center[ # ```r g + theme_minimal() ``` <!-- --> ```r g + theme_dark() ``` <!-- --> ]] --- class: split-20 .row[.content[ #Even More Pre-Defined Themes ```r library(ggthemes) ``` ]] .row[ .split-two[ .column[.content.center[ ```r g + theme_stata() + scale_color_stata() ``` <!-- --> ]] .column[.content.center[ ```r g + theme_solarized() + scale_color_solarized() ``` 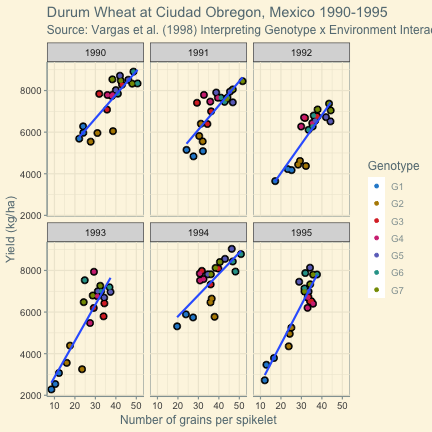<!-- --> ]] ]] --- class: split-30 .column.bg-blue[.content[ # Cheatsheet * There are even more features in `ggplot2` and many extension packages. * Go to RStudio > Help > Cheatsheets > Data Visualisation with ggplot2. * There is a useful addin for modifying the theme: `ggThemeAssist`. ]] <div class="column" style="background-image: url('images/ggplot-cheatsheet.png'); background-size: contain; background-repeat: no-repeat"> </div> --- class: split-two .column.bg-yellow[.content[ # References * Wickham, Hadley (2016) ggplot2: Elegant Graphics for Data Analysis. Springer *2nd Edition* * https://ggplot2.tidyverse.org/ # What graph to choose for your data? * Visit https://www.data-to-viz.com/ * Wilke, Clause (Pre-release) Fundamentals of Data Visualisation https://serialmentor.com/dataviz/ ]] .column.bg-main1.white[.content[ # How to get help There is a vibrant, friendly and (overly)-generous community of users of R (which is another reason that makes using R great). * [RStudio Community](https://community.rstudio.com/) * [Stack Overflow](https://stackoverflow.com/questions/tagged/ggplot2?sort=frequent&pageSize=50) * [R for Data Scence Online Learning Community Slack Channel](https://www.jessemaegan.com/post/r4ds-the-next-iteration/) * [DataCamp Courses](https://www.datacamp.com/courses/data-visualization-with-ggplot2-1) (only first chapter is free) * Ask on [twitter using #rstats](https://twitter.com/search?q=%23rstats&src=typd&lang=en) (better to go above forums instead) ]] --- class: bg-main1 split-30 hide-slide-number .column.bg-main3[ ] .column.slide-in-right[ .sliderbox.bg-main2.vmiddle[ # .font_large[`plotly`] ]] --- class: split-70 with-border .column[.content[ # `plotly` - interactive graphics ```r library(plotly) g <- ggplot(iris, aes(Sepal.Length, Sepal.Width, color=Species)) + geom_point() *ggplotly(g) ``` <center> <iframe src="iris_ggplotly.html" width="600px" height="500px" frameBorder="0"> </iframe> </center> ]] .column.bg-yellow[.content.center.vmiddle[ <img src="images/plotly-logo.png"> ]] --- class: split-70 with-border .column[.content[ # Simple animation with `plotly` ```r g <- ggplot(vargas.wheat1.traits, * aes(NGS, yield, frame=year)) + geom_point(aes(color=gen)) + geom_smooth(method="lm") ggplotly(g) ``` <center> <iframe src="wheat_ggplotly.html" width="600px" height="500px" frameBorder="0"> </iframe> </center> ]] .column.bg-yellow[.content.vmiddle.center[ # .font_large[.font_large[<span><i class="fab fa-r-project faa-vertical animated " style=" color:#4286f4;"></i></span>]] ]] --- class: bg-main1 split-30 hide-slide-number .column.bg-main3[ ] .column.slide-in-right[ .sliderbox.bg-main2.vmiddle[ # .font_large[`desplot`] ]] --- class: split-70 with-border .column[.content[ # Yield of Oats in a Split-Plot Experiment ```r str(yates.oats) # from agridat ``` ``` ## 'data.frame': 72 obs. of 8 variables: ## $ row : int 16 12 3 14 8 5 15 11 3 14 ... ## $ col : int 3 4 3 1 2 2 4 4 4 2 ... ## $ yield: int 80 60 89 117 64 70 82 102 82 114 ... ## $ nitro: num 0 0 0 0 0 0 0.2 0.2 0.2 0.2 ... ## $ gen : Factor w/ 3 levels "GoldenRain","Marvellous",..: 1 1 1 1 1 1 1 1 1 1 ... ## $ block: Factor w/ 6 levels "B1","B2","B3",..: 1 2 3 4 5 6 1 2 3 4 ... ## $ grain: num 20 15 22.2 29.2 16 ... ## $ straw: num 28 25 40.5 28.8 32 ... ``` .bottom_abs.width100.font_small[ Yates, Frank (1935) Complex experiments. *Journal of the Royal Statistical Society Suppl* **2** 181-247 ] ]] .column.bg-green[.content.vmiddle[ <img src="images/oats.jpg" width="114%" style="margin-right: -7%; margin-left: -7%;"> ]] --- class: split-70 with-border .column[.content[ # `desplot` - visualising designs for field trials ```r library(desplot) desplot(block ~ row + col, yates.oats, col=nitro, text=gen, cex=1, aspect=176/620, out1=block, out2=gen, out2.gpar=list(col = "gray50", lwd = 1, lty = 1)) ``` <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-107-1.png" style="display: block; margin: auto;" /> ]] .column.bg-green[.content.vmiddle.center[ <img src="images/desplot-logo.png" width="60%"> ]] --- class: split-70 with-border .column[.content[ # `ggplot` version of `desplot` in beta now! Enable by using argument `gg=TRUE` in `desplot`. <img src="day1-session02-datavis_files/figure-html/unnamed-chunk-108-1.png" style="display: block; margin: auto;" /> This means that you can use `theme` and other `ggplot` features easily. ]] .column.bg-green[.content.vmiddle.center[ <img src="images/desplot-logo.png" width="60%"> <img src="images/ggplot2.png" width="60%"> ]] --- class: split-40 title-slide2 with-border white background-image: url("images/bg1.jpg") background-size: cover .column[.content[ ]] .column.shade_main[.content[ <br><br> # <u>Slides</u> These slides were made using the R package [`xaringan`](https://github.com/yihui/xaringan) with the [`ninja-themes`](https://github.com/emitanaka/ninja-theme) and is available at [`bit.ly/UT-WS-DataVis`](http://bit.ly/UT-WS-DataVis). # <u>Your Turn</u> <s>Download `day1-session02-datavis-tutorial.Rmd` here, open in RStudio, push the button "Run Document" on the top tab and work through the exercises.</s> For workshop participants, contact Emi for the tutorials. <br><br> ### <a rel='license' href='http://creativecommons.org/licenses/by-sa/4.0/'><img alt='Creative Commons License' style='border-width:0; width:30pt' src='images/cc.svg' /><img alt='Creative Commons License' style='border-width:0; width:20pt' src='images/by.svg' /><img alt='Creative Commons License' style='border-width:0; width:20pt' src='images/sa.svg' /></a><span style='font-size:10pt'> This work by <span xmlns:cc='http://creativecommons.org/ns#' property='cc:attributionName'>Emi Tanaka</span> is licensed under a <a rel='license' href='http://creativecommons.org/licenses/by-sa/4.0/'>Creative Commons Attribution-ShareAlike 4.0 International License</a>.</span> ]] <img src="images/USydLogo-white.svg" style="position:absolute; top:80%; left:80%;width:200px">