ETC3250/5250
Introduction to Machine Learning
Neural network II
Lecturer: Emi Tanaka
Department of Econometrics and Business Statistics
Acknowledgement
This lecture benefited from the lecture notes by Dr. Ruben Loaiza-Maya.
Wide and deep neural networks
Neural network layers
- The neural network we saw last week had 3 layers:
- Input layer (2 nodes)
- Middle or hidden layer (3 nodes)
- Output layer (3 nodes)
- The number of nodes in the input and output layer are constrained by the input data and desired output (regression or classification).
Wide neural network
- We refer to neural network with large number of neurons (nodes) in the hidden layer as a wide neural network.
- The number of parameters in the neural network model generally increases exponentially as the number of neurons increase.
- Wide neural networks are however harder to calibrate as the number of neurons increase.
Here, the addition of 2 neurons increases the number of parameters by 10.
Deep neural network
- Deep neural networks, also called feed forward neural networks, add more layers.
- The number of parameters also can increase exponentially.
- But it can achieve flexibility with less neuron and it can be faster to calibrate.
Feed forward neural networks
Mathematical notation
- We now use a new mathematical notation for neural networks:
- L denotes the total number of layers,
- a^{(l)}_k denotes the value for layer l for k-th node,
- K_l denotes the total number of nodes in layer l,
- h_l denotes the activation function in layer l,
- b_k^{(l)} denotes the bias in node k of layer l,
- \boldsymbol{w}_k^{(l)} denotes the weights for k-th node of layer l.
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x})
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x})
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
- \boldsymbol{a}^{(2)} = (a^{(2)}_1, a^{(2)}_2, a^{(2)}_3)^\top
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
- \boldsymbol{a}^{(2)} = (a^{(2)}_1, a^{(2)}_2, a^{(2)}_3)^\top
Layer 3
- The activation function h_3 is Tanh.
- a^{(3)}_1 = h_2(b_1^{(3)}+\boldsymbol{w}_1^{(3)\top}\boldsymbol{a}^{(2)})
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
- \boldsymbol{a}^{(2)} = (a^{(2)}_1, a^{(2)}_2, a^{(2)}_3)^\top
Layer 3
- The activation function h_3 is Tanh.
- a^{(3)}_1 = h_3(b_1^{(3)}+\boldsymbol{w}_1^{(3)\top}\boldsymbol{a}^{(2)}), a^{(3)}_2 = h_3(b_2^{(3)}+\boldsymbol{w}_2^{(3)\top}\boldsymbol{a}^{(2)})
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
- \boldsymbol{a}^{(2)} = (a^{(2)}_1, a^{(2)}_2, a^{(2)}_3)^\top
Layer 3
- The activation function h_3 is Tanh.
- a^{(3)}_1 = h_3(b_1^{(3)}+\boldsymbol{w}_1^{(3)\top}\boldsymbol{a}^{(2)}), a^{(3)}_2 = h_3(b_2^{(3)}+\boldsymbol{w}_2^{(3)\top}\boldsymbol{a}^{(2)})
- \boldsymbol{a}^{(3)} = (a^{(3)}_1, a^{(3)}_2)^\top
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
- \boldsymbol{a}^{(2)} = (a^{(2)}_1, a^{(2)}_2, a^{(2)}_3)^\top
Layer 3
- The activation function h_3 is Tanh.
- a^{(3)}_1 = h_3(b_1^{(3)}+\boldsymbol{w}_1^{(3)\top}\boldsymbol{a}^{(2)}), a^{(3)}_2 = h_3(b_2^{(3)}+\boldsymbol{w}_2^{(3)\top}\boldsymbol{a}^{(2)})
- \boldsymbol{a}^{(3)} = (a^{(3)}_1, a^{(3)}_2)^\top
Output layer
- The activation function h_4 is Softmax.
- f_1(\boldsymbol{x}) = h_4(b_1^{(4)}+\boldsymbol{w}_1^{(4)\top}\boldsymbol{a}^{(3)})
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
- \boldsymbol{a}^{(2)} = (a^{(2)}_1, a^{(2)}_2, a^{(2)}_3)^\top
Layer 3
- The activation function h_3 is Tanh.
- a^{(3)}_1 = h_3(b_1^{(3)}+\boldsymbol{w}_1^{(3)\top}\boldsymbol{a}^{(2)}), a^{(3)}_2 = h_3(b_2^{(3)}+\boldsymbol{w}_2^{(3)\top}\boldsymbol{a}^{(2)})
- \boldsymbol{a}^{(3)} = (a^{(3)}_1, a^{(3)}_2)^\top
Output layer
- The activation function h_4 is Softmax.
- f_1(\boldsymbol{x}) = h_4(b_1^{(4)}+\boldsymbol{w}_1^{(4)\top}\boldsymbol{a}^{(3)}), f_2(\boldsymbol{x}) = h_4(b_2^{(4)}+\boldsymbol{w}_2^{(4)\top}\boldsymbol{a}^{(3)})
Forward evaluation
Input layer
\boldsymbol{x} = (1, x_1)^\top
Layer 2
- The activation function h_2 is ReLU.
- a^{(2)}_1 = h_2(\boldsymbol{\beta}_1^\top\boldsymbol{x}), a^{(2)}_2 = h_2(\boldsymbol{\beta}_2^\top\boldsymbol{x}), a^{(2)}_3 = h_2(\boldsymbol{\beta}_3^\top\boldsymbol{x})
- \boldsymbol{a}^{(2)} = (a^{(2)}_1, a^{(2)}_2, a^{(2)}_3)^\top
Layer 3
- The activation function h_3 is Tanh.
- a^{(3)}_1 = h_3(b_1^{(3)}+\boldsymbol{w}_1^{(3)\top}\boldsymbol{a}^{(2)}), a^{(3)}_2 = h_3(b_2^{(3)}+\boldsymbol{w}_2^{(3)\top}\boldsymbol{a}^{(2)})
- \boldsymbol{a}^{(3)} = (a^{(3)}_1, a^{(3)}_2)^\top
Output layer
- The activation function h_4 is Softmax.
- f_1(\boldsymbol{x}) = h_4(b_1^{(4)}+\boldsymbol{w}_1^{(4)\top}\boldsymbol{a}^{(3)}), f_2(\boldsymbol{x}) = h_4(b_2^{(4)}+\boldsymbol{w}_2^{(4)\top}\boldsymbol{a}^{(3)}), f_3(\boldsymbol{x}) = h_4(b_3^{(4)}+\boldsymbol{w}_3^{(4)\top}\boldsymbol{a}^{(3)})
General feed forward neural network
- Input layer: \boldsymbol{x} = (1, x_1, \dots, x_p)^\top
- Layer 2: \boldsymbol{a}^{(2)} = (h_2(\boldsymbol{\beta}^\top_1\boldsymbol{x}), \dots, h_2(\boldsymbol{\beta}^\top_{K_2}\boldsymbol{x}))^\top
- Layer 3: \boldsymbol{a}^{(3)} = (h_3(b_1^{(3)}+\boldsymbol{w}^{(3)\top}_1\boldsymbol{a}^{(2)}), \dots, h_3(b_{K_3}^{(3)}+\boldsymbol{w}^{(3)\top}_{K_3}\boldsymbol{a}^{(2)}))^\top
- \cdots
- Layer l:\boldsymbol{a}^{(l)} = (h_l(b_1^{(l)}+\boldsymbol{w}^{(l)\top}_1\boldsymbol{a}^{(l - 1)}), \dots, h_l(b_{K_l}^{(l)}+\boldsymbol{w}^{(l)\top}_{K_l}\boldsymbol{a}^{(l-1)}))^\top
- \cdots
- Layer L -1: \boldsymbol{a}^{(L-1)} = (h_{L-1}(b_1^{(L-1)}+\boldsymbol{w}^{(L-1)\top}_1\boldsymbol{a}^{(L-2)}), \dots, h_{L-1}(b_{K_{L-1}}^{({L-1})}+\boldsymbol{w}^{({L-1})\top}_{K_{L-1}}\boldsymbol{a}^{(L-2)}))^\top
- Output layer: f_k(\boldsymbol{x}) = h_L(b^{(L)}+\boldsymbol{w}_k^{(L)\top}\boldsymbol{a}^{(L-1)}).
- The output actually depends on the parameters: \boldsymbol{\theta} = (\underbrace{\beta_1, \dots, \beta_{K_2}}_{\text{coefficients}}, \underbrace{b_1^{(3)}, \dots, b_{K_{L-1}}^{(L-1)}}_{\text{biases}},\underbrace{\boldsymbol{w}_1^{(3)\top}, \dots, \boldsymbol{w}_k^{(L)\top}}_{\text{weights}})^\top.
- We can write this dependency more explicitly as f_k(\boldsymbol{x}~|~\boldsymbol{\theta}).
- How do we calibrate (or train) these parameters \boldsymbol{\theta}?
Calibration
Calibration
- As like other models, we:
- define a loss function, and
- find parameters that minimise this loss function.
- What loss function we use depends on the problem.
Regression loss
- Find \boldsymbol{\theta} that minimises the mean squared error:
\text{MSE}(\boldsymbol{\theta}) = \frac{1}{n}\sum_{i=1}^n\left(y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta})\right)^2
Binary classification loss
- Find \boldsymbol{\theta} that minimises the binary cross-entropy (BCE):
\text{BCE}(\boldsymbol{\theta}) = -\frac{1}{n}\sum_{i=1}^n\left\{y_i\log(P(y_i=1~|~\boldsymbol{x}_i,\boldsymbol{\theta})) + (1-y_i)\log(1 - P(y_i=1~|~\boldsymbol{x}_i,\boldsymbol{\theta}))\right\}
Multi-class classification loss
- Find \boldsymbol{\theta} that minimises the cross-entropy (CE):
\text{CE}(\boldsymbol{\theta}) = -\frac{1}{n}\sum_{i=1}^n\sum_{j=1}^my_{ij}\log(P(y_{ij} = 1~|~\boldsymbol{x}_i,\boldsymbol{\theta}))
Optimisation
- Regardless of whether it is regression or classification, we must find hyperparameters that optimise a loss function.
- Finding these hyperparameters is often hard with no closed-form solution.
- There are various optimisation methods to find these hyperparameters – we discuss:
- gradient descent, and
- stochastic gradient descent.
Illustrative example
- Suppose that we have a regression problem that require callibration of one hyperparameter with MSE loss function.
- Our goal is to find \theta that corresponds to the bottom of the curve.
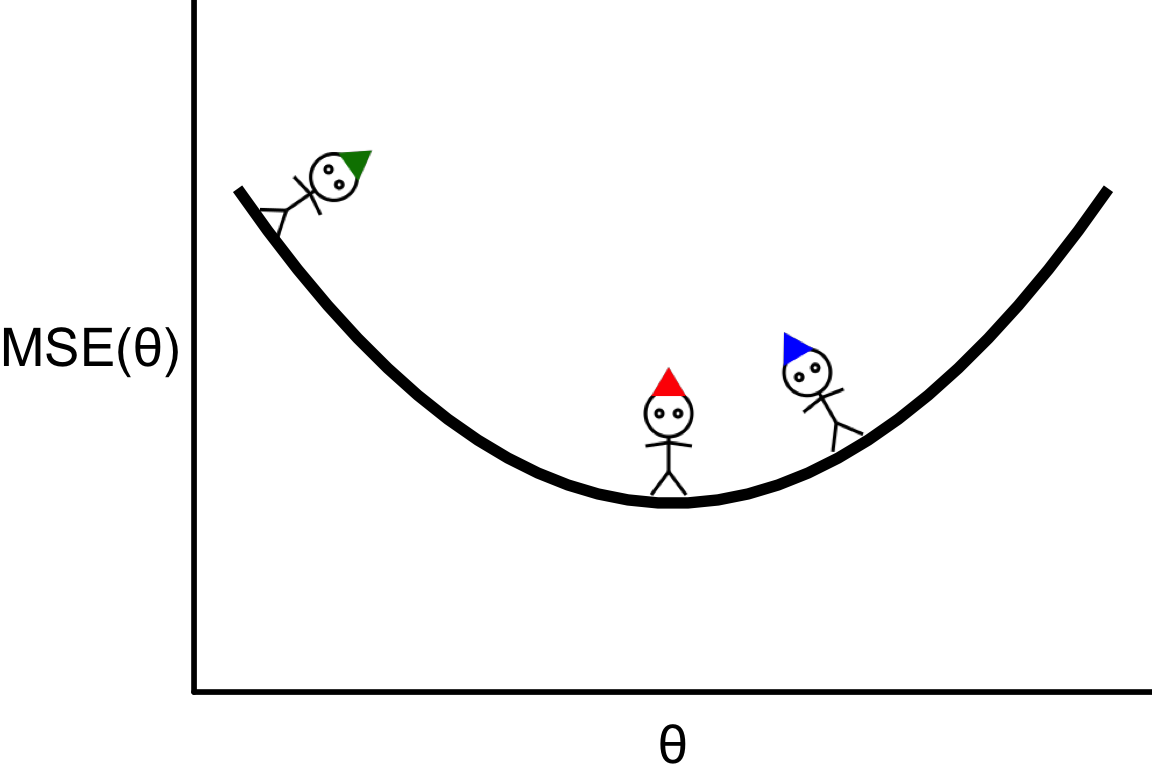
Finding the zero slope
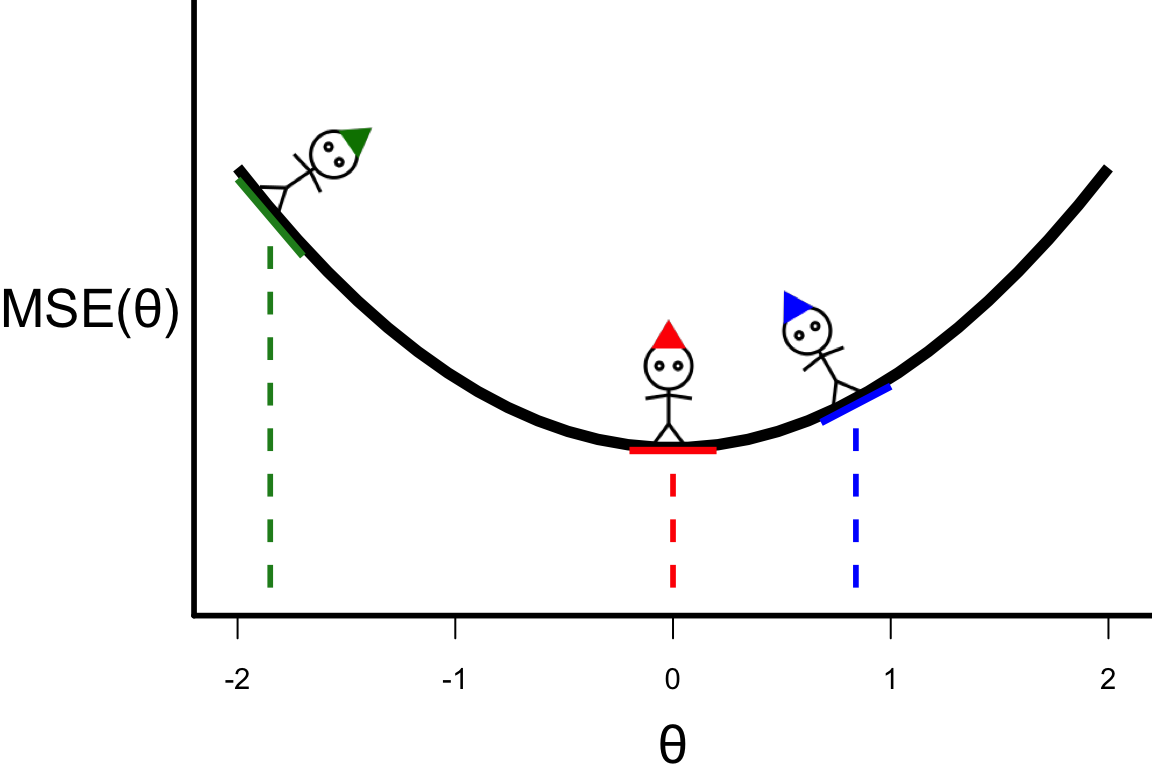
- Suppose that we calculate \text{MSE}(\theta) for three values of \theta.
- For this function, the optimal \theta is where the individual with the red hat is.
- This is also where the slope (or derivative) is zero: \frac{\partial \text{MSE}(\theta)}{\partial \theta} = 0.
Optimising a multi-variable function
- In practice, \boldsymbol{\theta} is typically a vector of length d with d > 1.
- The optimal value of \boldsymbol{\theta} is then found by solving for \boldsymbol{\theta} such that \nabla \text{MSE}(\boldsymbol{\theta}) = 0 where \nabla \text{MSE}(\boldsymbol{\theta}) = \left(\frac{\partial\text{MSE}(\boldsymbol{\theta})}{\partial\theta_1}, \dots, \frac{\partial\text{MSE}(\boldsymbol{\theta})}{\partial\theta_d}\right)^\top is the vector of slopes for all parameters (gradient).
- Finding \boldsymbol{\theta} such that \nabla \text{MSE}(\boldsymbol{\theta}) = 0 is hard!
- We use gradient descent to find this.
Searching for the optimal value
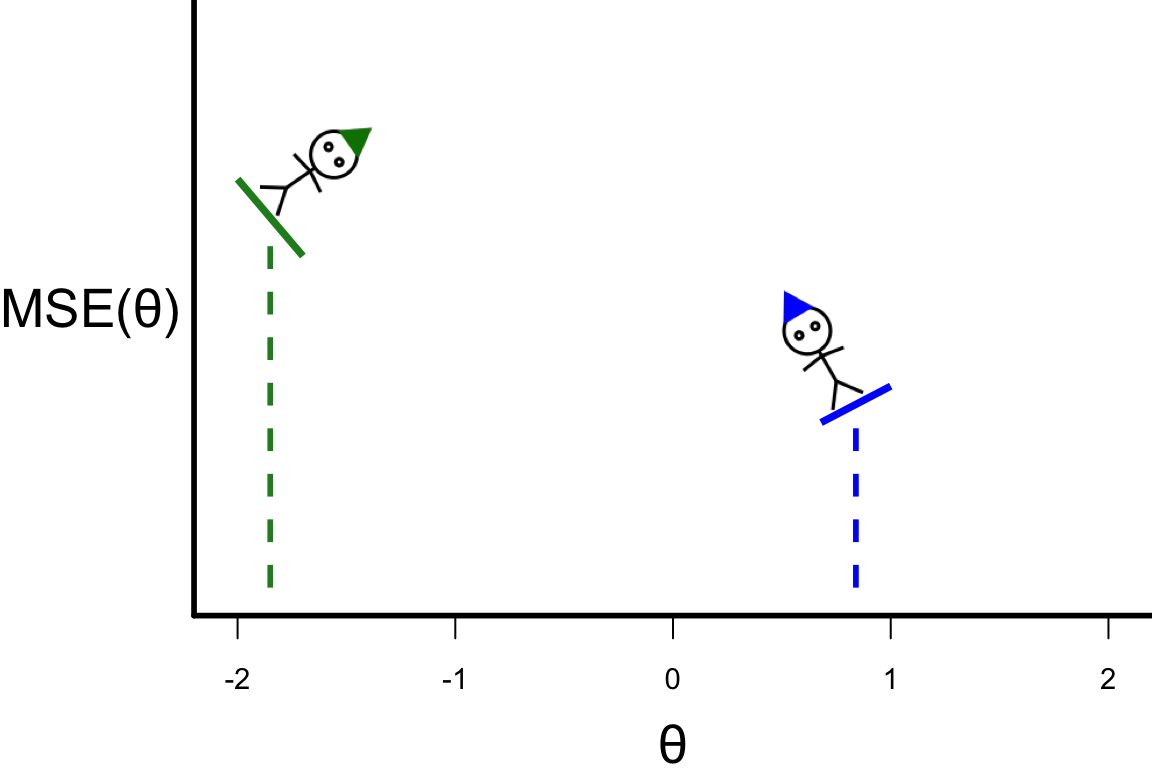
- To search for the optimal value of \theta, we choose some starting points.
- We can calculate the slope at the starting points giving us a guide where to search next: \theta^{[s+1]} = \theta^{[s]} - r \left.\frac{\partial\text{MSE}(\theta)}{\partial\theta}\right\vert_{\theta=\theta^{[s]}}, where r > 0 is the length of the step.
Illustrative example: Step 0
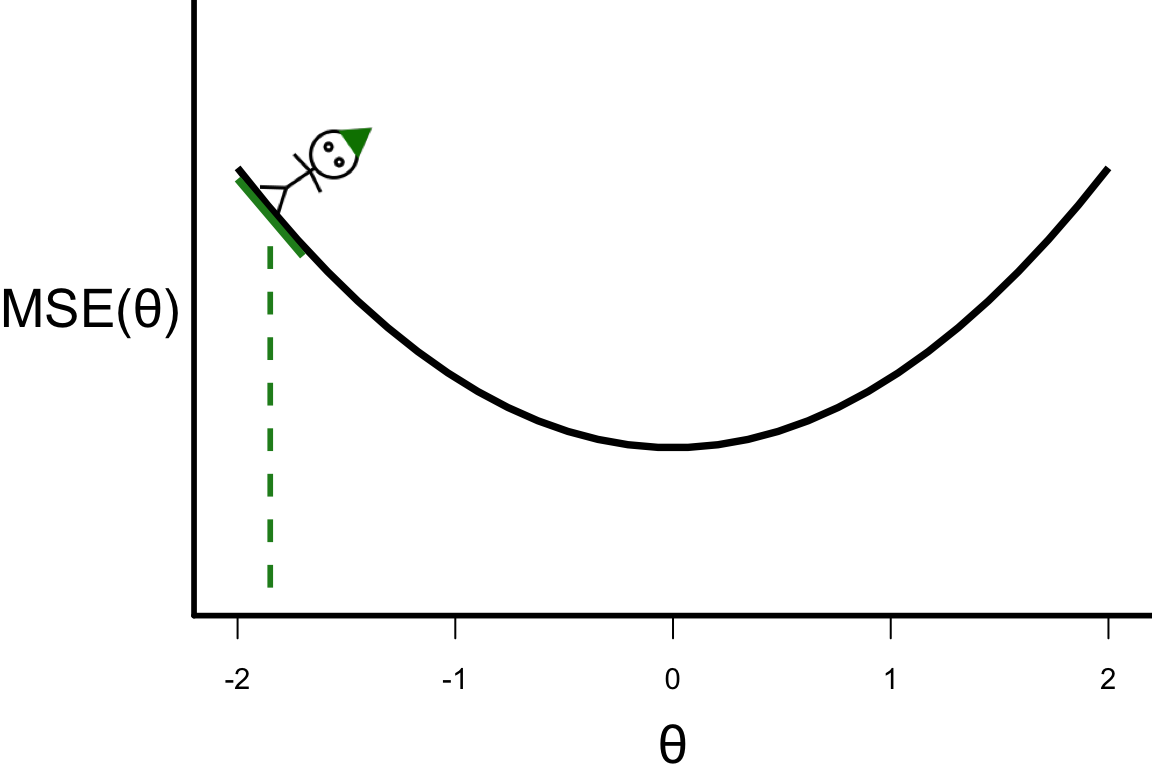
\theta^{[s+1]} = \theta^{[s]} - r \left.\frac{\partial\text{MSE}(\theta)}{\partial\theta}\right\vert_{\theta=\theta^{[s]}}
- Suppose
- \frac{\partial\text{MSE}(\theta)}{\partial\theta} = 4\theta,
- r = 0.1, and
- the starting point \theta^{[0]} = -1.8.
- \theta^{[1]} = -1.8 - 0.1 \times 4 \times (-1.8) = -1.08.
Illustrative example: Step 1
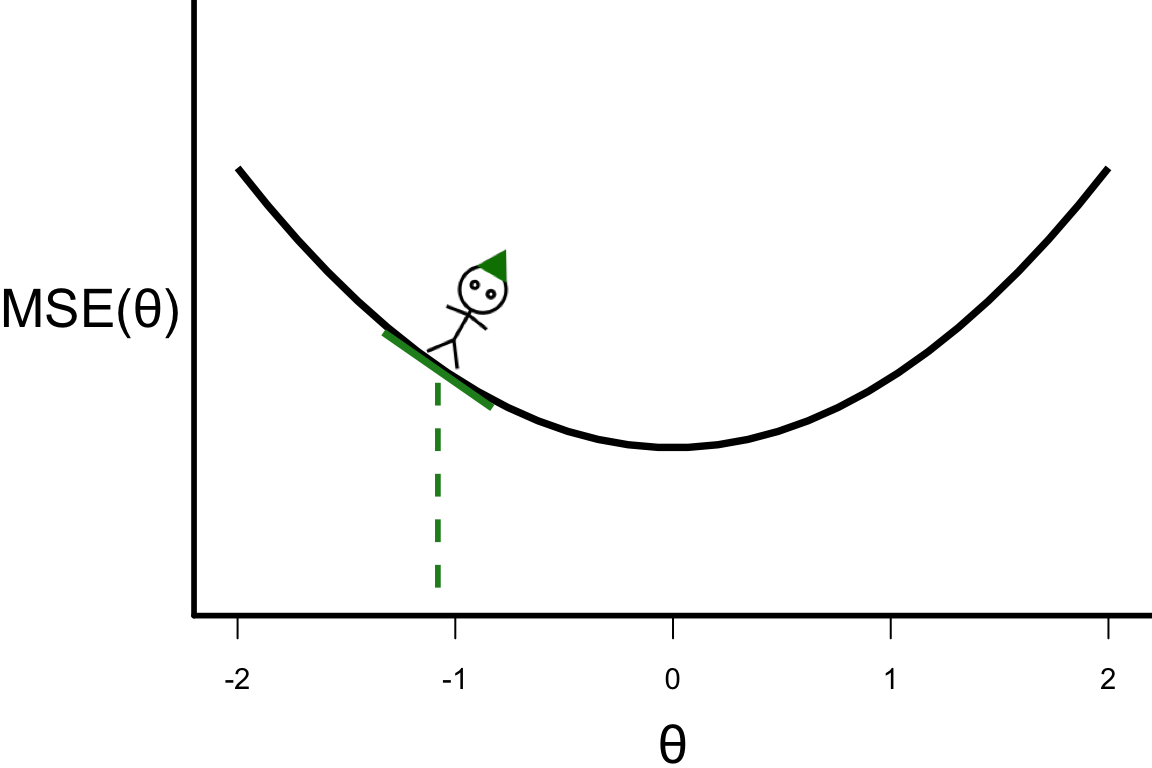
\theta^{[s+1]} = \theta^{[s]} - r \left.\frac{\partial\text{MSE}(\theta)}{\partial\theta}\right\vert_{\theta=\theta^{[s]}}
- \theta^{[1]} = -1.08
- \theta^{[2]} = -1.08 - 0.1 \times 4 \times (-1.08) = -0.648.
Illustrative example: Step 2
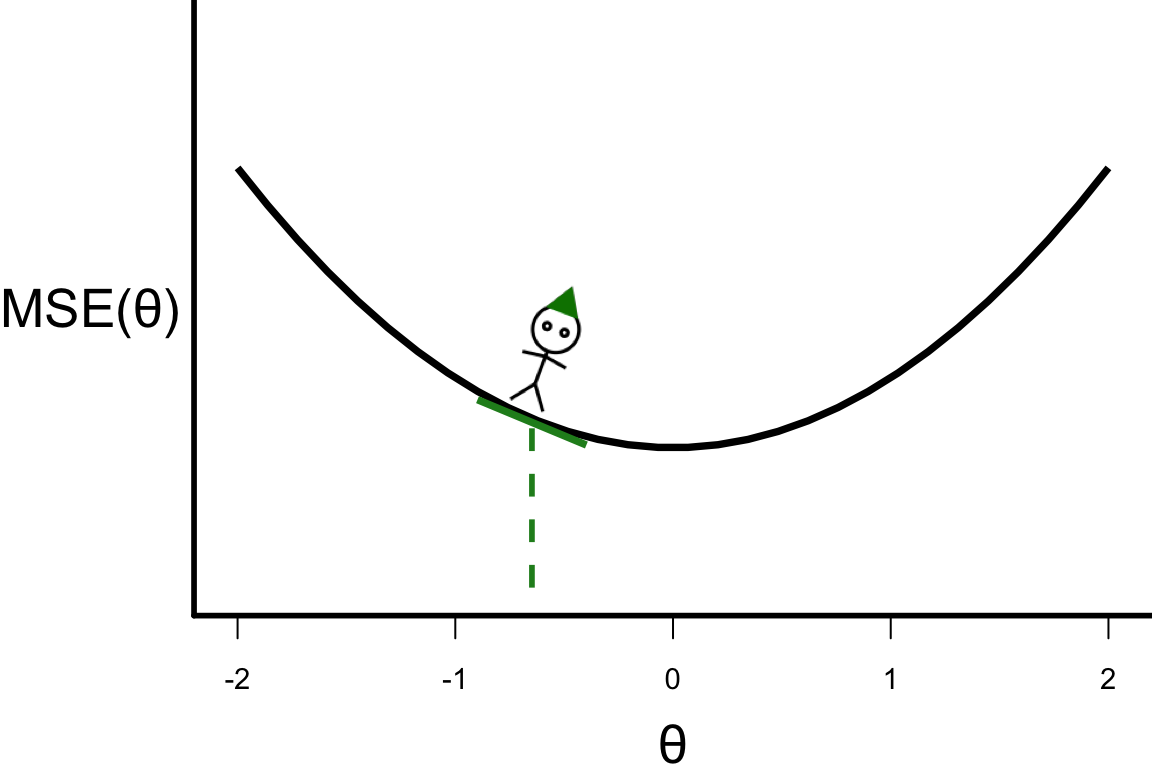
\theta^{[s+1]} = \theta^{[s]} - r \left.\frac{\partial\text{MSE}(\theta)}{\partial\theta}\right\vert_{\theta=\theta^{[s]}}
- \theta^{[2]} = -0.648
- \theta^{[3]} = -0.648 - 0.1 \times 4 \times (-0.648) = -0.3888.
Illustrative example: Step 3
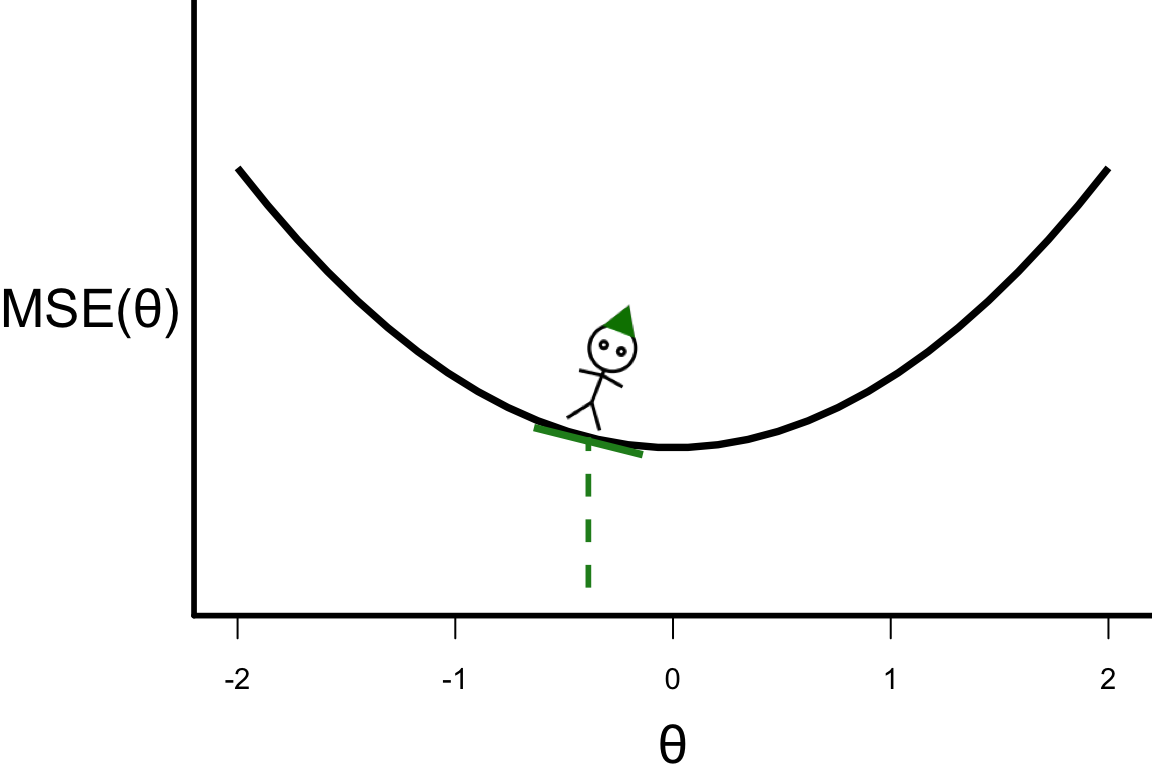
\theta^{[s+1]} = \theta^{[s]} - r \left.\frac{\partial\text{MSE}(\theta)}{\partial\theta}\right\vert_{\theta=\theta^{[s]}}
- \theta^{[3]} = -0.3888
- \theta^{[4]} = -0.3888 - 0.1 \times 4 \times (-0.3888) = -0.23328
Illustrative example: Step 4
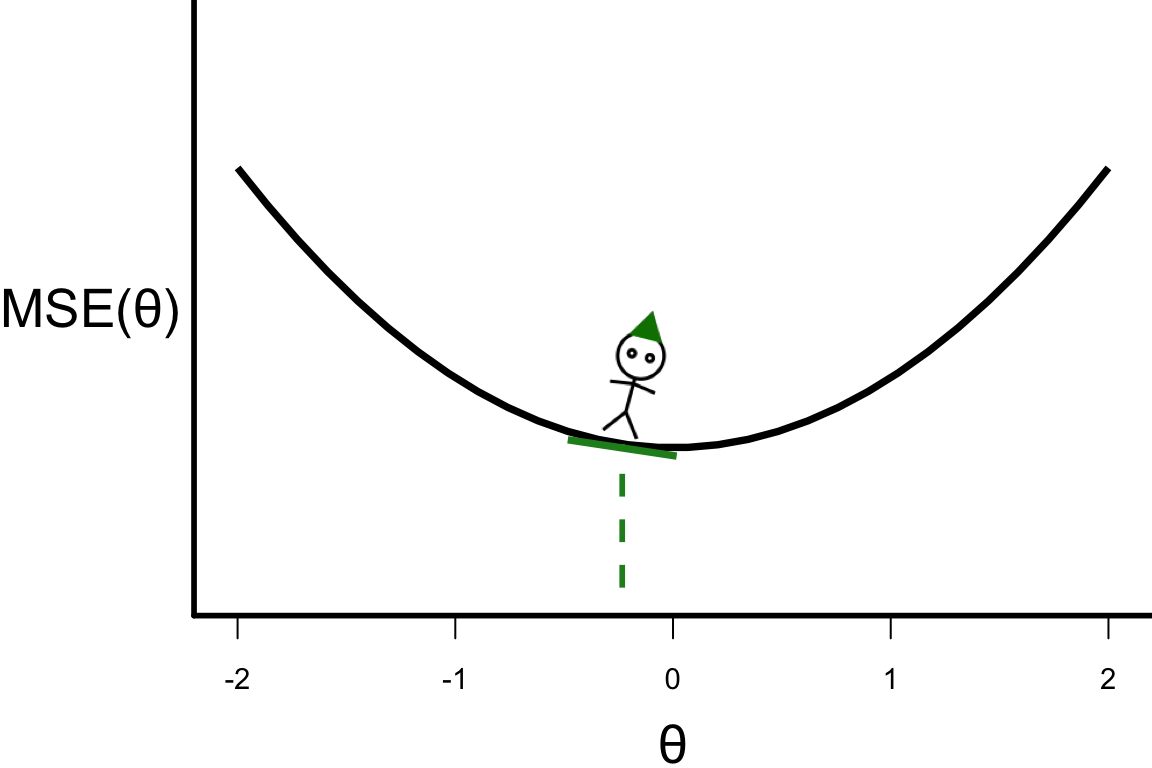
\theta^{[s+1]} = \theta^{[s]} - r \left.\frac{\partial\text{MSE}(\theta)}{\partial\theta}\right\vert_{\theta=\theta^{[s]}}
- \theta^{[4]} = -0.23328
- \theta^{[5]} = -0.23328 - 0.1 \times 4 \times (-0.23328) = -0.139968
\cdots
Illustrative example: Step 20
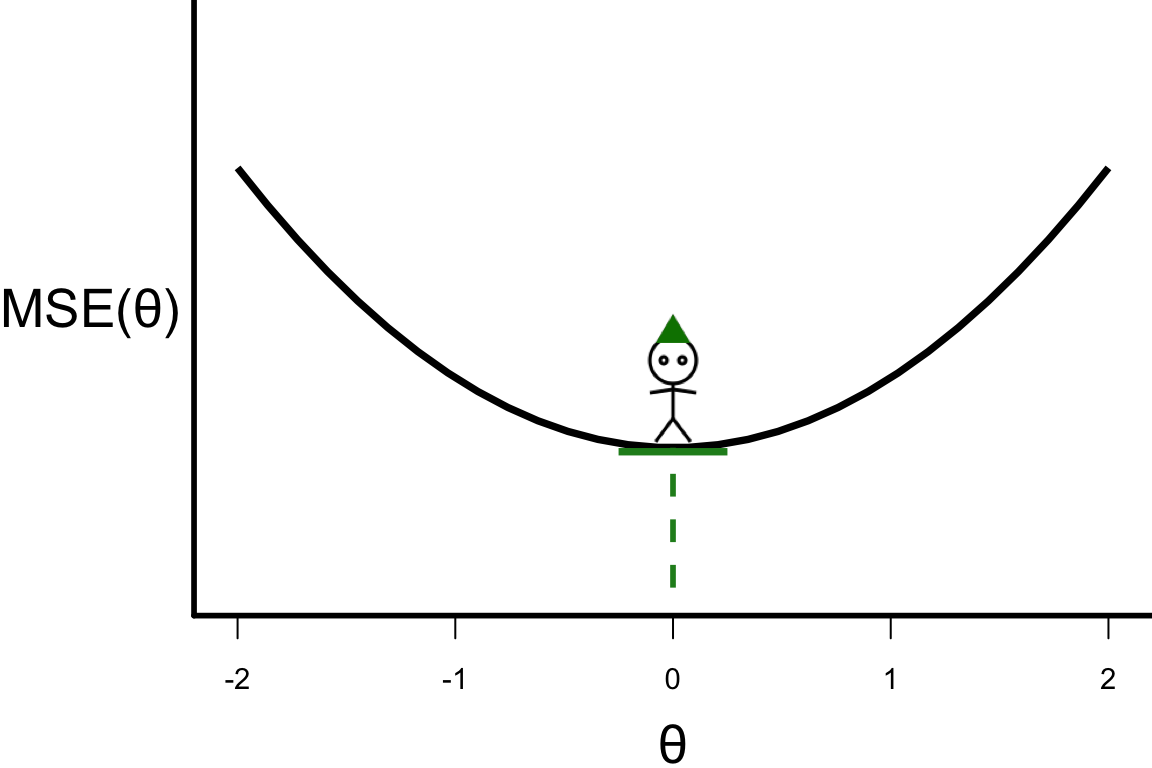
\theta^{[s+1]} = \theta^{[s]} - r \left.\frac{\partial\text{MSE}(\theta)}{\partial\theta}\right\vert_{\theta=\theta^{[s]}}
- \theta^{[20]} = -0.00006581085
- What if there are more than one parameter?
Gradient descent
- In the multi-variable version: \boldsymbol{\theta}^{[s+1]} = \left.\boldsymbol{\theta}^{[s]} - r\nabla \text{MSE}(\boldsymbol{\theta})\right\vert_{\theta=\theta^{[s]}}. where r>0 is the learning rate.
- You need a starting value \boldsymbol{\theta}^{[0]} and take a number of steps.
- This is known as the gradient descent method.
Limitations of gradient descent
- In practice, the loss function generally involves valuation for each observation, e.g. for MSE: \nabla \text{MSE}(\boldsymbol{\theta}) = \frac{1}{n}\sum_{i=1}^n\nabla\left(y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta})\right)^2.
- This can be computationally expensive if n is large.
- Gradient descent optimisation can get stuck in local optima.
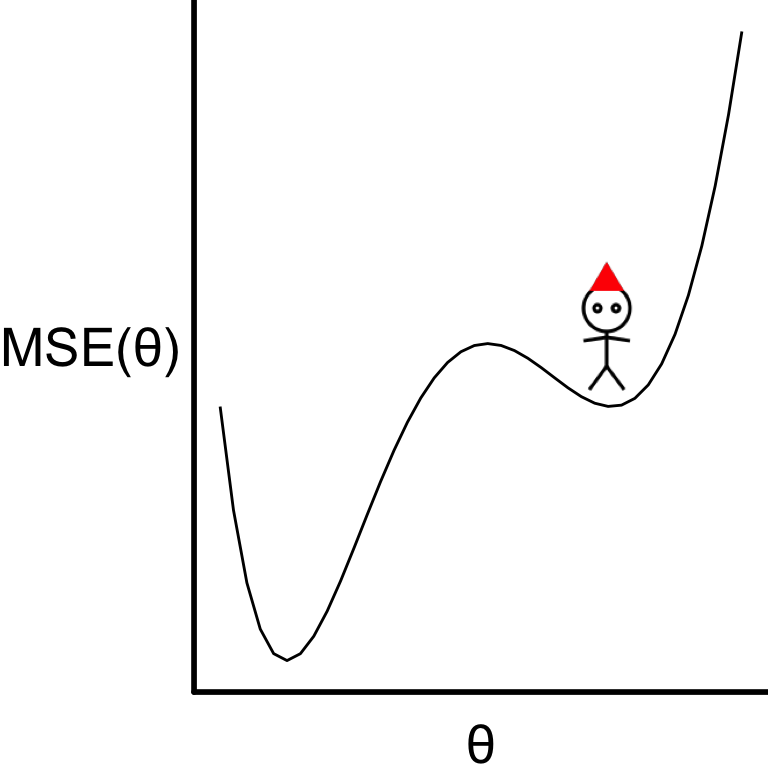
Stochastic gradient descent
Stochastic gradient descent
- In a stochastic gradient descent (SGD) considers at each step \boldsymbol{\theta}^{[s+1]} = \boldsymbol{\theta}^{[s]} - r \left.\nabla \hat{\text{MSE}}(\boldsymbol{\theta})\right\vert_{\boldsymbol{\theta}=\boldsymbol{\theta}^{[s]}} where \left.\nabla \hat{\text{MSE}}(\boldsymbol{\theta})\right\vert_{\boldsymbol{\theta}=\boldsymbol{\theta}^{[s]}}=\frac{1}{M}\sum_{i\in B^{[s]}}\nabla\left(y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta}^{[s]})\right)^2. where B^{[s]} denotes a set of random batch of M \ll n observations at iteration s.
Batch illustration
- Suppose we have n = 1000.
- We draw a (random without replacement) batch of size 3, say resulting in B^{[1]} = \{133, 606, 851\}.
- With gradient descent, \nabla\text{MSE}(\boldsymbol{\theta}) = \frac{1}{1000}\left(\nabla(y_1 - f(\boldsymbol{x}_1~|~\boldsymbol{\theta})^2 + \nabla(y_2 - f(\boldsymbol{x}_2~|~\boldsymbol{\theta})^2 + \cdots + \nabla(y_{1000} - f(\boldsymbol{x}_{1000}~|~\boldsymbol{\theta})^2\right).
- But for stochastic gradient descent, we replace this with: \nabla\hat{\text{MSE}}(\boldsymbol{\theta}) = \frac{1}{3}\left(\nabla(y_{133} - f(\boldsymbol{x}_{133}~|~\boldsymbol{\theta})^2 + \nabla(y_{606} - f(\boldsymbol{x}_{606}~|~\boldsymbol{\theta})^2 + \nabla(y_{851} - f(\boldsymbol{x}_{851}~|~\boldsymbol{\theta})^2\right).
- The latter is faster to calculate and can escape from local optima.
Epoch
- The number of epochs is the number of times that the algorithm has used all observations in the data.
- Illustrative data: y_1, y_2, y_3, y_4, y_5, y_6, y_7, y_8, y_9, y_{10}, y_{11}, y_{12}
Epoch
- The number of epochs is the number of times that the algorithm has used all observations in the data.
- Illustrative data: y_1, y_2, y_3, y_4, y_5, y_6, y_7, y_8, y_9, y_{10}, y_{11}, y_{12}
- s = 1, B^{[1]} = \color{red}{\{4, 8, 10\}}
Epoch
- The number of epochs is the number of times that the algorithm has used all observations in the data.
- Illustrative data: y_1, y_2, y_3, y_4, y_5, y_6, y_7, y_8, y_9, y_{10}, y_{11}, y_{12}
- s = 1, B^{[1]} = \{4, 8, 10\}
- s = 2, B^{[2]} = \color{red}{\{1, 5, 12\}}
Epoch
- The number of epochs is the number of times that the algorithm has used all observations in the data.
- Illustrative data: y_1, y_2, y_3, y_4, y_5, y_6, y_7, y_8, y_9, y_{10}, y_{11}, y_{12}
- s = 1, B^{[1]} = \{4, 8, 10\}
- s = 2, B^{[2]} = \{1, 5, 12\}
- s = 3, B^{[3]} = \color{red}{\{2, 3, 6\}}
Epoch
- The number of epochs is the number of times that the algorithm has used all observations in the data.
- Illustrative data: y_1, y_2, y_3, y_4, y_5, y_6, y_7, y_8, y_9, y_{10}, y_{11} , y_{12}
- s = 1, B^{[1]} = \{4, 8, 10\}
- s = 2, B^{[2]} = \{1, 5, 12\}
- s = 3, B^{[3]} = \{2, 3, 6\}
- s = 4, B^{[4]} = \color{red}{\{7, 9, 11\}}
Epoch
- The number of epochs is the number of times that the algorithm has used all observations in the data.
- Illustrative data: y_1, y_2, y_3, y_4, y_5, y_6, y_7, y_8, y_9, y_{10}, y_{11} , y_{12}
- s = 1, B^{[1]} = \{4, 8, 10\}
- s = 2, B^{[2]} = \{1, 5, 12\}
- s = 3, B^{[3]} = \{2, 3, 6\}
- s = 4, B^{[4]} = \{7, 9, 11\}
Epoch 1
Epoch
- The number of epochs is the number of times that the algorithm has used all observations in the data.
- Illustrative data: y_1, y_2, y_3, y_4, y_5, y_6, y_7, y_8, y_9, y_{10}, y_{11} , y_{12}
- s = 1, B^{[1]} = \{4, 8, 10\}
- s = 2, B^{[2]} = \{1, 5, 12\}
- s = 3, B^{[3]} = \{2, 3, 6\}
- s = 4, B^{[4]} = \{7, 9, 11\}
Epoch 1
- s = 5, B^{[5]} = \{3, 4, 11\}
- s = 6, B^{[6]} = \{2, 7, 9\}
- s = 7, B^{[7]} = \{6, 10, 12\}
- s = 8, B^{[8]} = \{1, 5, 8\}
Epoch 2
Backpropagation
In evaluating \nabla \hat{\text{MSE}}(\boldsymbol{\theta}), we must evaluate \nabla (y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta}))^2.
Now we have: \nabla (y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta}))^2 = - 2(y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta}))\times \underbrace{\frac{\partial f(\boldsymbol{x}_i|\boldsymbol{\theta})}{\partial \boldsymbol{a}_i^{(L - 1)}}}_{\text{Layer }L -1}\times \underbrace{\frac{\partial \boldsymbol{a}_i^{(L - 1)}}{\partial \boldsymbol{a}_i^{(L - 2)}}}_{\text{Layer }L -2}\times \underbrace{\frac{\partial \boldsymbol{a}_i^{(L - 2)}}{\partial \boldsymbol{a}_i^{(L - 3)}}}_{\text{Layer }L -3}\times \cdots \times \underbrace{\frac{\partial \boldsymbol{a}_i^{(2)}}{\partial \boldsymbol{\theta}}}_{\text{Layer }1}.
The gradient of the later layers is calculated earlier.
For parameter \theta, we can show \frac{\partial (y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta}))^2}{\partial \theta} = \text{constant}\times \underbrace{\frac{\partial h_L(a_i^{(L-1)})}{\partial a_i^{(L-1)}}}_{\text{Layer }L -1}\times \underbrace{\frac{\partial h_L(a_i^{(L-2)})}{\partial a_i^{(L-2)}}}_{\text{Layer }L -2}\times \underbrace{\frac{\partial h_L(a_i^{(L-3)})}{\partial a_i^{(L-3)}}}_{\text{Layer }L -3}\times \cdots \times \underbrace{\frac{\partial h_2(x_i| \theta)}{\partial \theta}}_{\text{Layer }1}.
Vanishing gradient problem
\frac{\partial (y_i - f(\boldsymbol{x}_i~|~\boldsymbol{\theta}))^2}{\partial \theta} = \text{constant}\times \underbrace{\frac{\partial h_L(a_i^{(L-1)})}{\partial a_i^{(L-1)}}}_{\text{Layer }L -1}\times \underbrace{\frac{\partial h_L(a_i^{(L-2)})}{\partial a_i^{(L-2)}}}_{\text{Layer }L -2}\times \underbrace{\frac{\partial h_L(a_i^{(L-3)})}{\partial a_i^{(L-3)}}}_{\text{Layer }L -3}\times \cdots \times \underbrace{\frac{\partial h_2(x_i| \theta)}{\partial \theta}}_{\text{Layer }1}.
- If h_l is the Sigmoid activation function then 0 < \frac{\partial h_l(a_i^{(l -1)})}{\partial a_i^{(l -1)}} < 1.
- If the L is large then this can result in \nabla (y_i - f(\boldsymbol{x}_i | \boldsymbol{\theta}))^2 \approx 0.
- This phenomenon is referred to as the vanishing gradient problem and results in difficulty with calibrating the neural network.
- To avoid this problem, you can use a different activation function that does not have the same issue, e.g. ReLU.
An application to fashion MNIST data
Fashion MNIST data
scroll
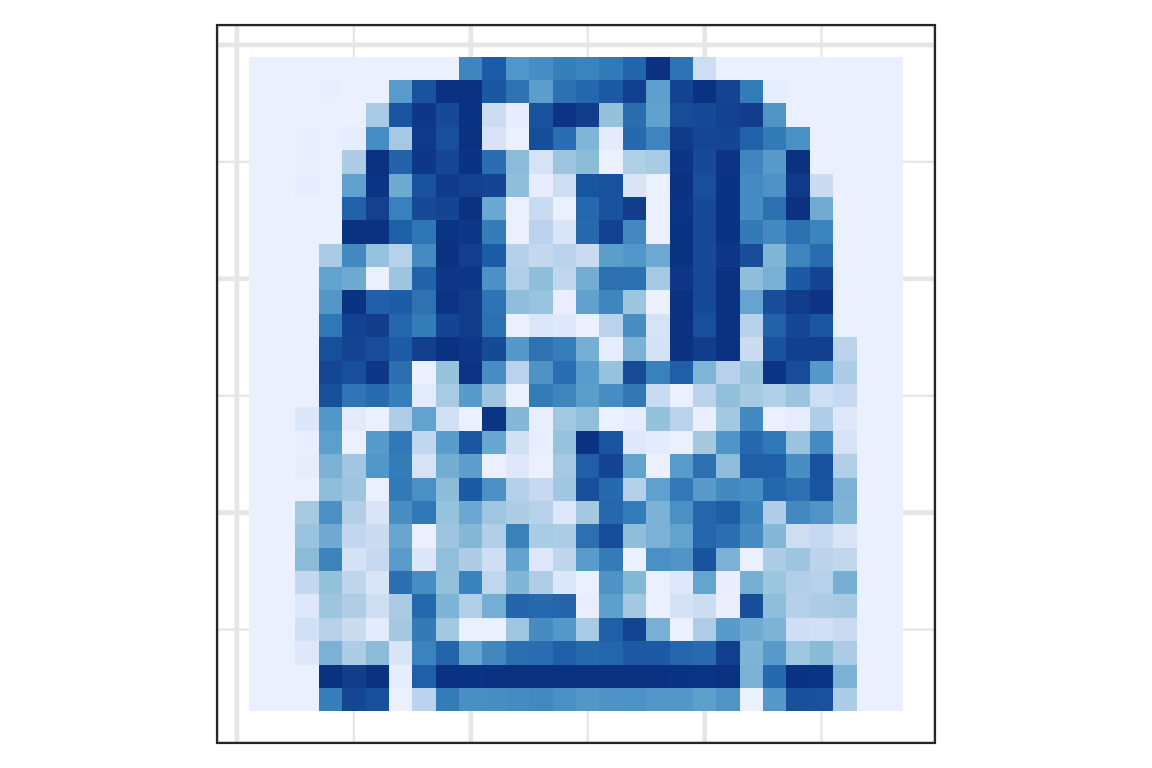
- The fashion MNIST data by Zalando SE contains 28 \times 28 pixel images of fashion items labelled as:
| Label | Description |
|---|---|
| 0 | T-shirt/top |
| 1 | Trouser |
| 2 | Pullover |
| 3 | Dress |
| 4 | Coat |
| 5 | Sandal |
| 6 | Shirt |
| 7 | Sneaker |
| 8 | Bag |
| 9 | Ankle boot |
- So this is a multi-class classification problem with m = 10 classes (or levels).
- The training data contains 60,000 observations with 784 variables (
pixel1, …,pixel784) and 1 response variable (label) labelled from 0-9. - The testing data contains 10,000 observations (note: this testing data also contains
label!).
Neural network in R 1 prepare the data
- To fit a neural network model, we use the
keraspackage.
- We first need to prepare the data:
- convert categorical data to dummy variables, and
- normalise the predictors.
Neural network in R 2 Define architechture
library(keras)
NN <- keras_model_sequential() %>%
# hidden layer
layer_dense(units = 128, # number of nodes in hidden layer
activation = "relu",
# number of predictors
input_shape = 784) %>%
# output layer
layer_dense(units = 10, # the number of classes
# we need to use softmax for multi-class classification
activation = "softmax")
NNModel: "sequential"
________________________________________________________________________________
Layer (type) Output Shape Param #
================================================================================
dense_1 (Dense) (None, 128) 100480
dense (Dense) (None, 10) 1290
================================================================================
Total params: 101,770
Trainable params: 101,770
Non-trainable params: 0
________________________________________________________________________________Neural network in R 3 Choose loss function
- For regression, you can use
mean_squared_error. - For binary classification, you can use
binary_crossentropy. - For multi-class classfication, you can use
categorical_crossentropy. - For the full list of loss functions, see
help("loss-functions").
Neural network in R 4 Fit model
This model takes long to fit!
- Typically
epochsis a large number.
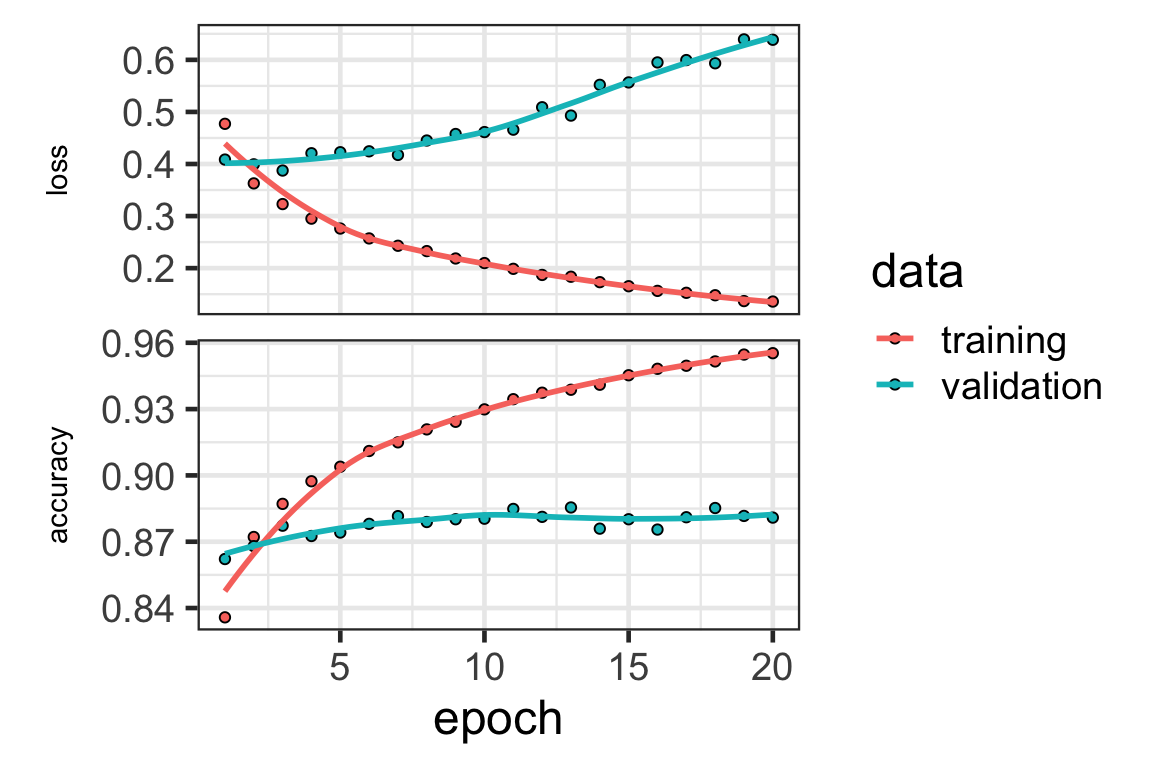
Model diagnostic
- We can plot the loss (and other metrics specified) of the neural network using the
plotfunction on the training history.
Predicting from neural network
- Notice we are using
NNland notlearnNNlhere!
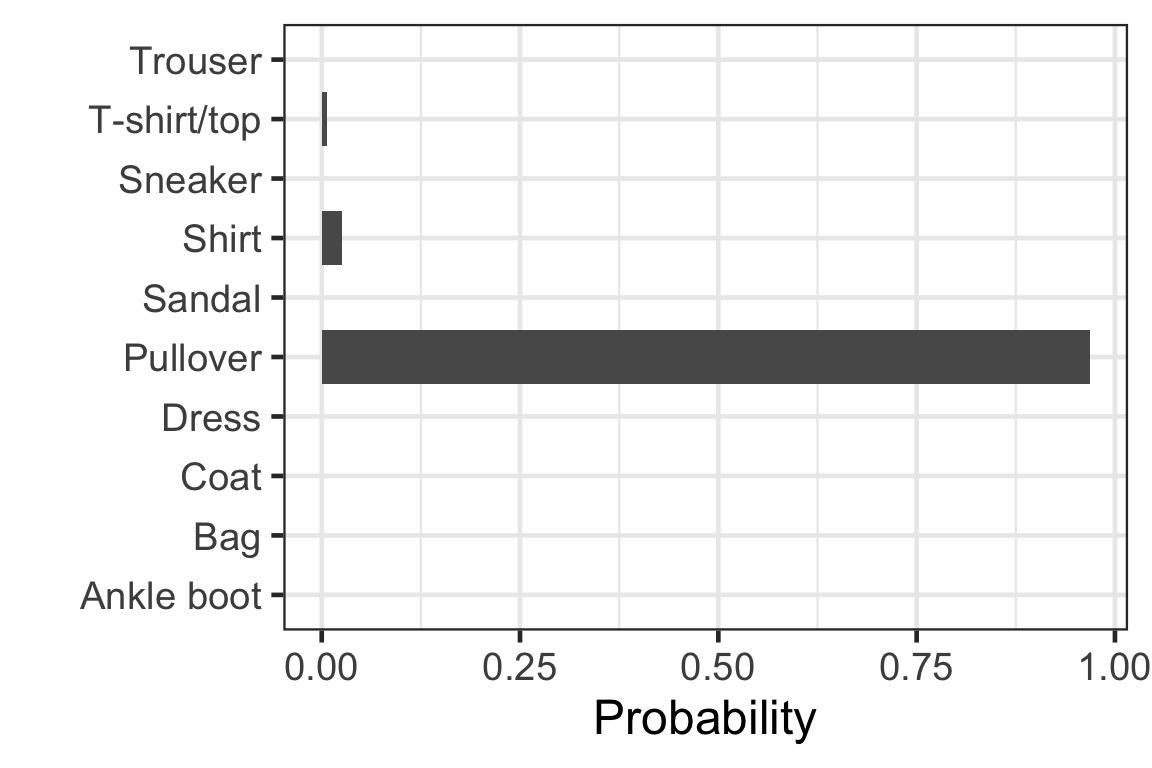
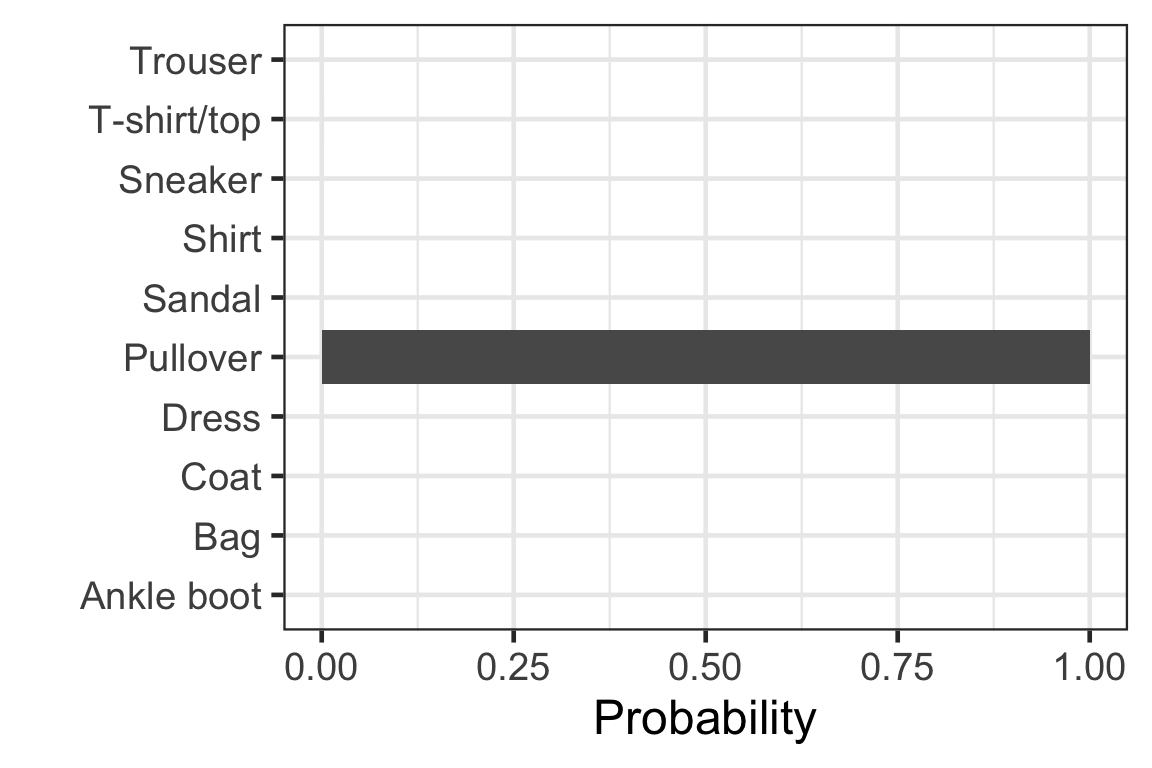
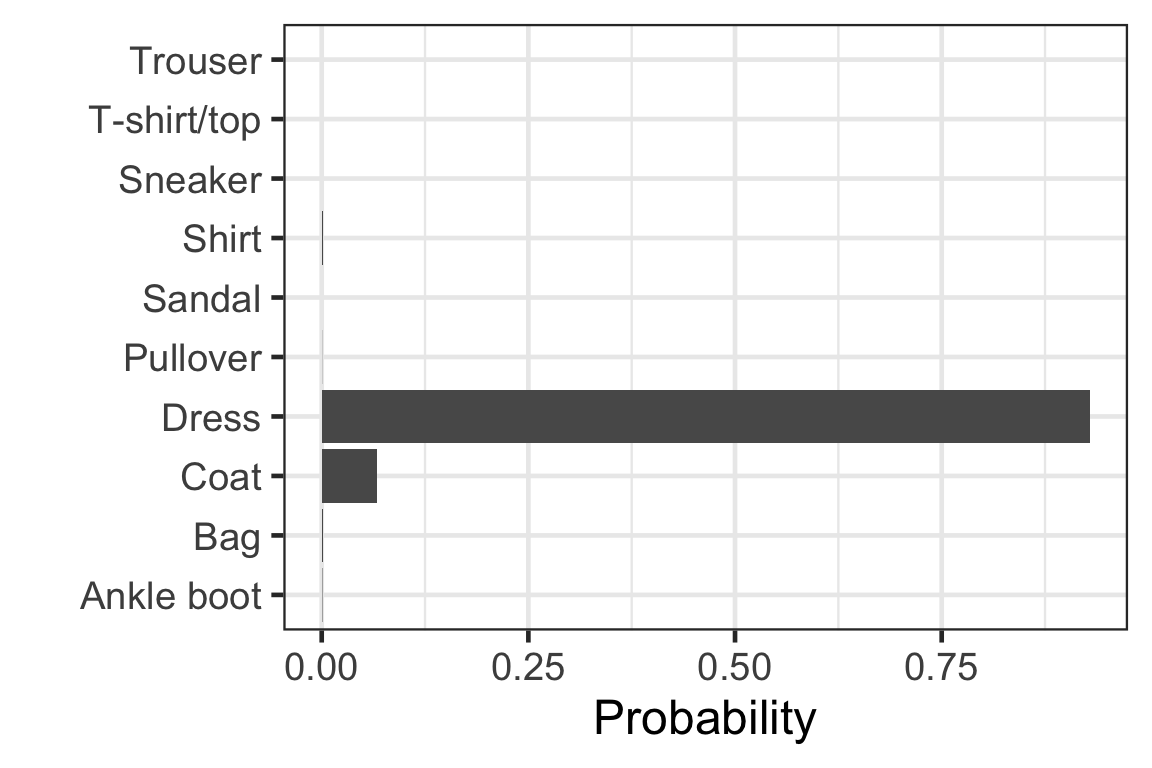
[,1] [,2] [,3] [,4] [,5]
[1,] 9.959991e-01 4.470216e-15 2.498414e-06 1.280446e-06 6.907984e-09
[2,] 2.188681e-21 1.000000e+00 5.769808e-32 9.360376e-28 1.490198e-26
[3,] 6.637161e-03 6.112183e-16 9.681513e-01 3.647272e-13 6.959190e-06
[,6] [,7] [,8] [,9] [,10]
[1,] 2.304835e-11 3.900097e-03 9.305310e-19 9.703544e-05 4.587546e-13
[2,] 0.000000e+00 8.628763e-31 0.000000e+00 4.123140e-29 1.746240e-37
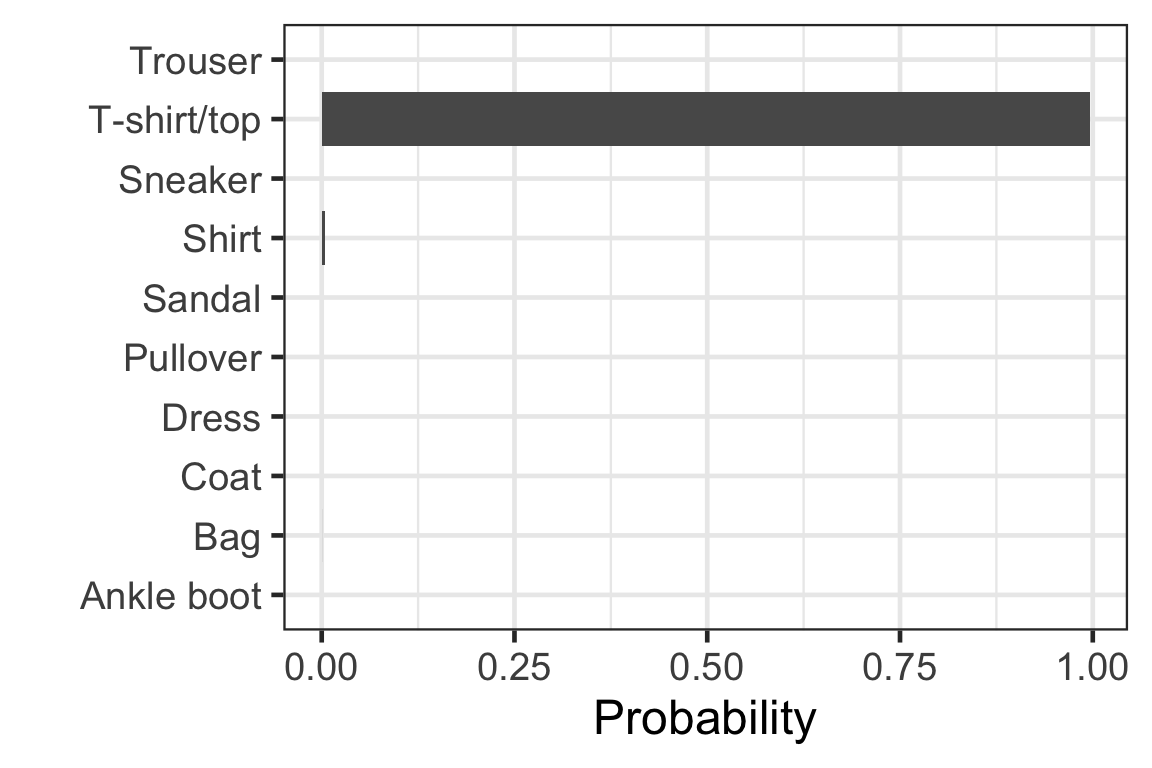
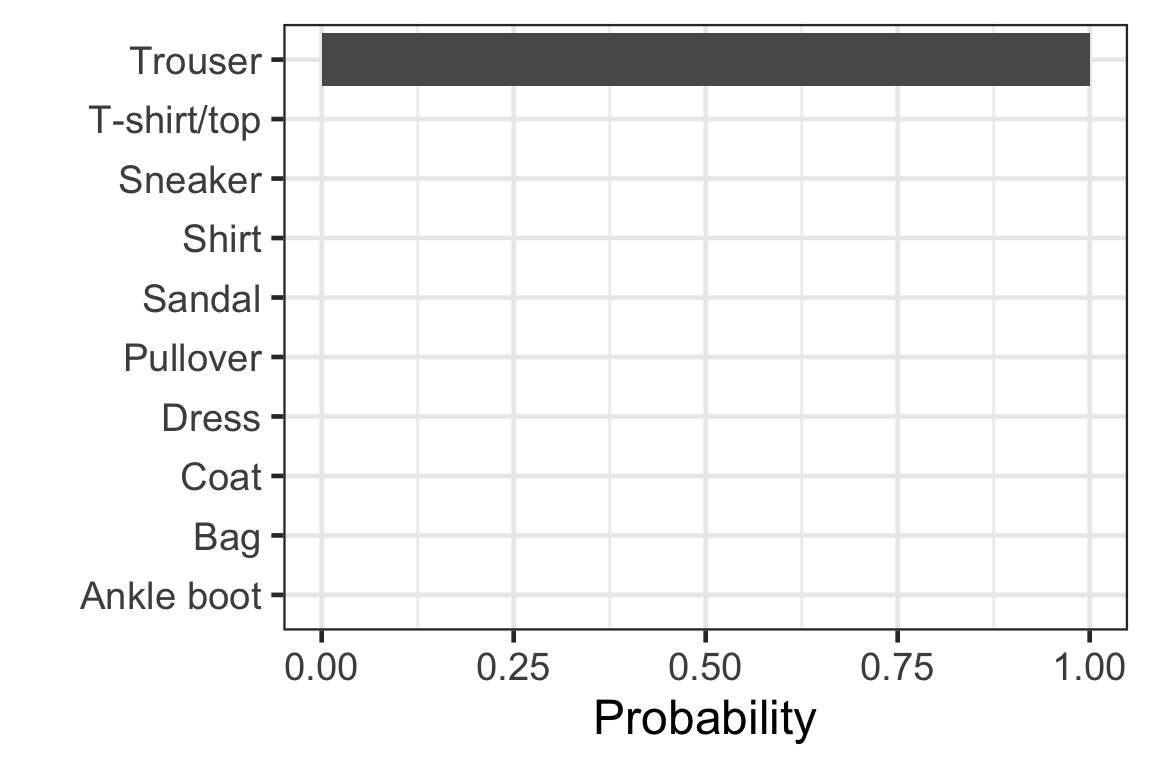
[3,] 8.466854e-11 2.520457e-02 8.891557e-21 6.381754e-13 2.705846e-17- Each row corresponds to the observation in the test data.
- Column c corresponds to the probability of the label c - 1.
- Sum of each row is 1.
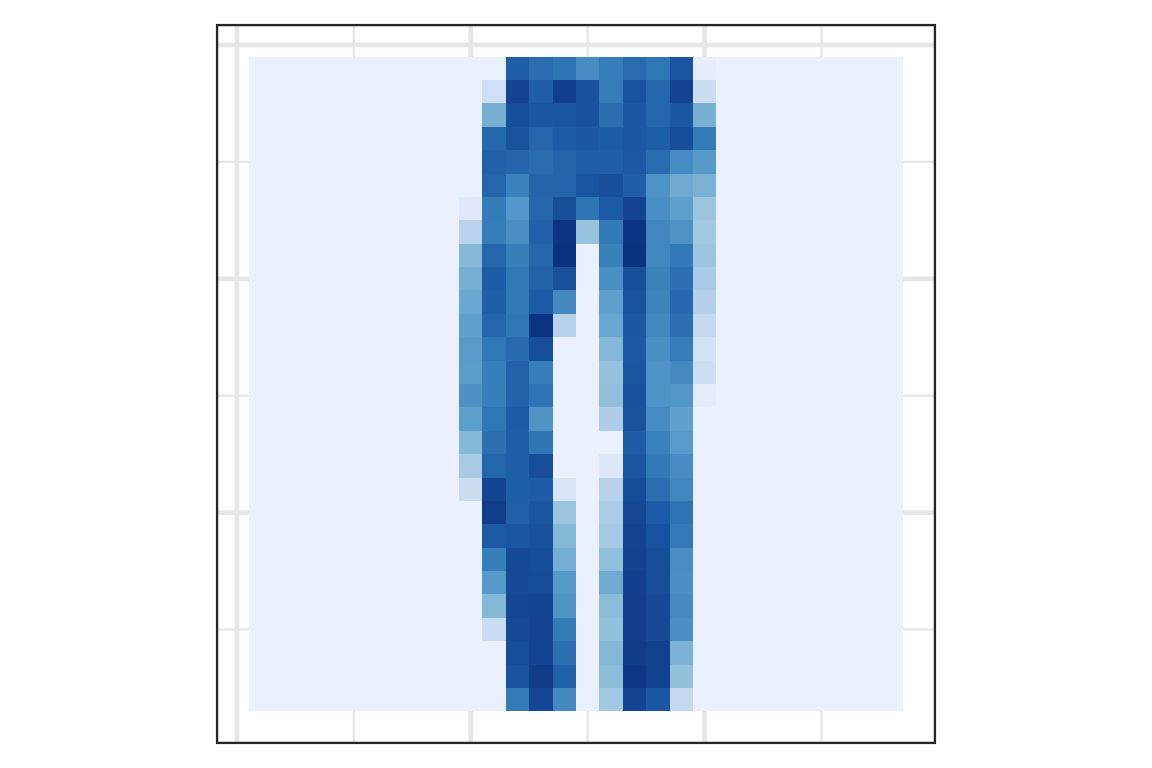
Visually checking the prediction: observation 1
Visually checking the prediction: observation 2
Visually checking the prediction: observation 3
Visually checking the prediction: observation 4
Visually checking the prediction: observation 5
Limitations
Cons of neural network
- Minimal interpretability.
- Requires the number of observations to be larger than the number of features.
- Computationally intensive
- Many calculations are required to estimate all of the parameters in many neural networks.
- Deep learning involves huge amounts of matrix multiplications and other operations.
- Often used in conjuction with GPUs to parallellise computations.
Takeaways
- To build a feed-forward neural network, we need the key components:
- Input data,
- A pre-defined network architecture,
- A feedback mechanism (e.g. loss function) to enable the network to learn, and
- Model training.
- Deep neural networks are faster to calibrate than wide neural networks via stochastic gradient descent and backpropogation algorithms.

ETC3250/5250 Week 12