Barplots and position adjustments in ggplot2
Data Visualisation with R
👩🏻💻 Emi Tanaka @ Monash University
- emi.tanaka@monash.edu
- @statsgen
- github.com/emitanaka
- emitanaka.org
28th November 2022 Australasian Applied Statistics Conference 2022
Position adjustments
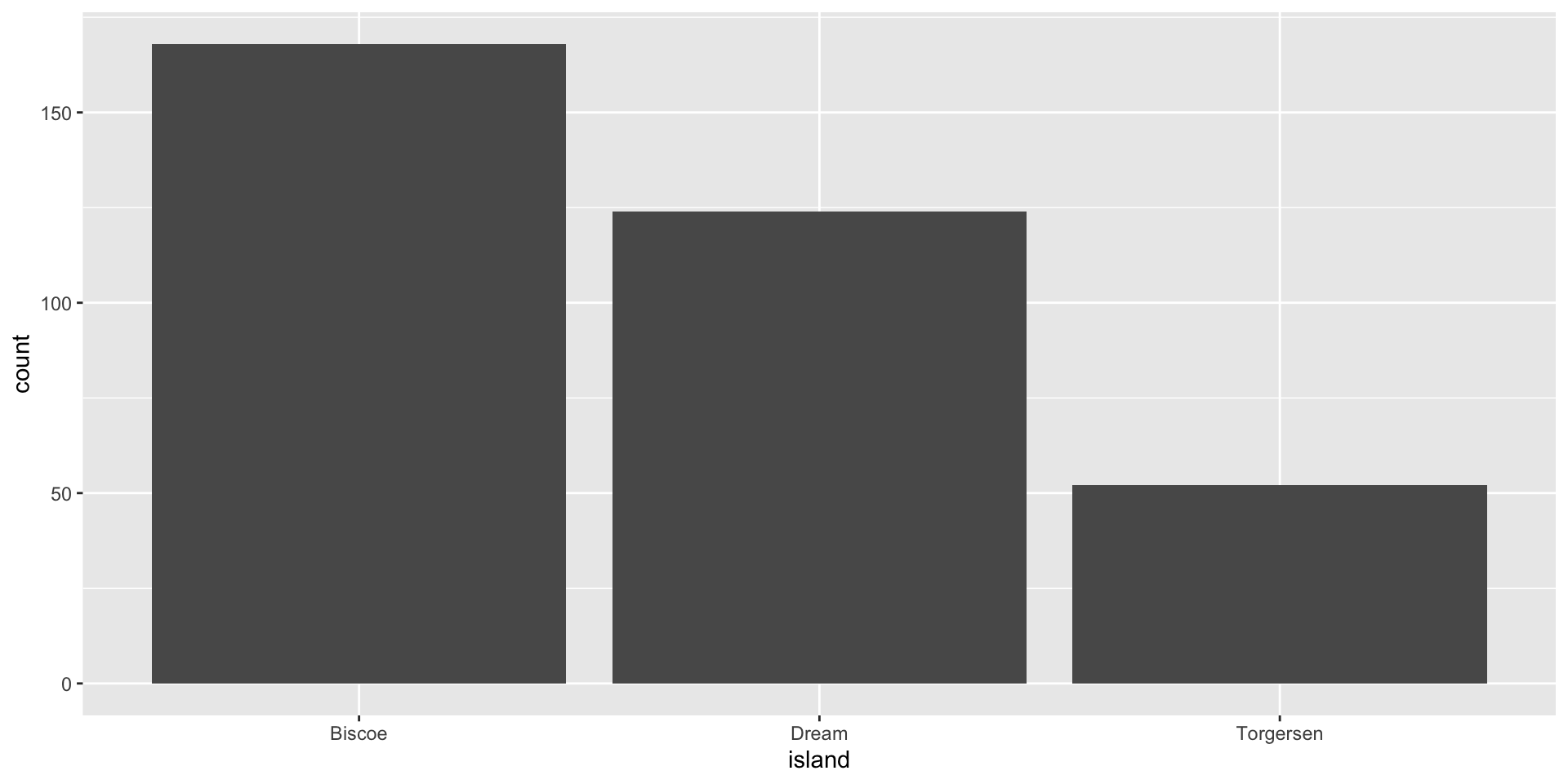
A barplot with geom_bar() with a categorical variable
- If you have a categorical variable, then you usually want to study the frequency of its categories.
- Here the
stat = "count"is computing the frequencies for each category for you.
A barplot with geom_bar() with a discrete numerical variable
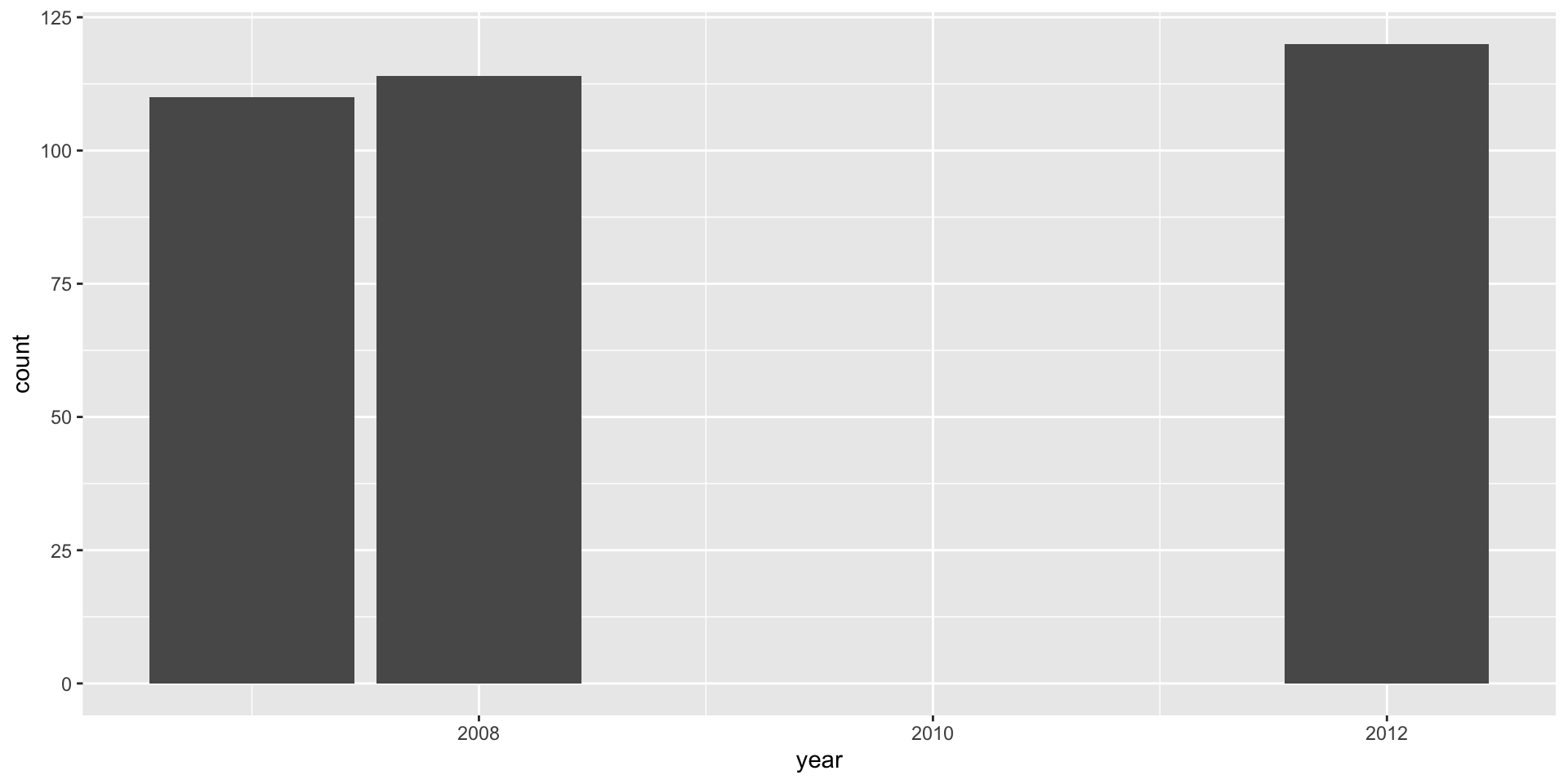
- If you supply a numerical variable, you can see now that the x-axis scale is continuous.
- If you want to study each level in a discrete variable, then you may want to convert the discrete variable to a factor instead
x = factor(year). - When the variable is a factor or character, the distances between the bars are equal and the labels correspond to that particular level.
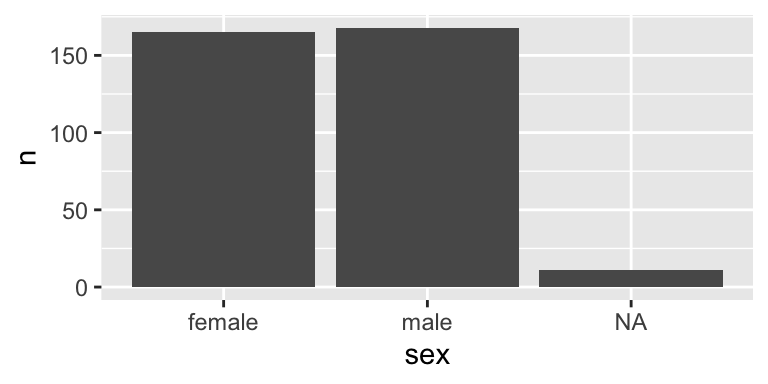
A barplot with geom_col()
- Sometimes your input data may already contain pre-computed counts.
# A tibble: 3 × 2
sex n
<fct> <int>
1 female 165
2 male 168
3 <NA> 11- In this case, you don’t need
stat = "count"to do the counting for you and usegeom_col()instead.
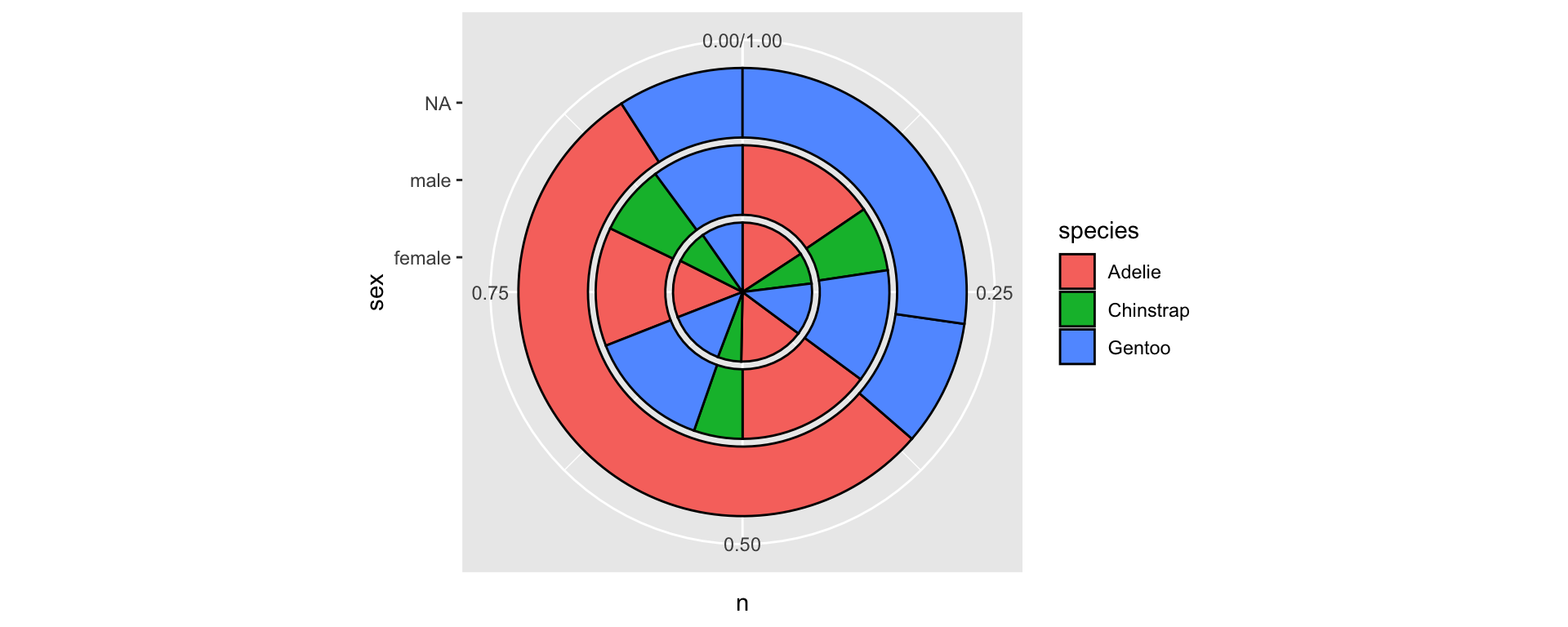
A stacked barplot with geom_col()
penguins %>%
group_by(species, sex, year) %>%
tally() %>%
ggplot(aes(year, n, fill = sex, group = year, color = species)) +
geom_col(position = "stack", linewidth = 8) +
geom_col(position = "stack", linewidth = 1, color = "black")
- By default the values in
yare stacked on top of another. - The aesthetic
grouphere breaks the count in two groups and stack one on top of the other (try running the code withoutgroup = year).
A grouped barplot with geom_col()
penguins %>%
group_by(sex, species, year) %>%
tally() %>%
ggplot(aes(sex, n, fill = species)) +
geom_col(color = "black", position = "dodge")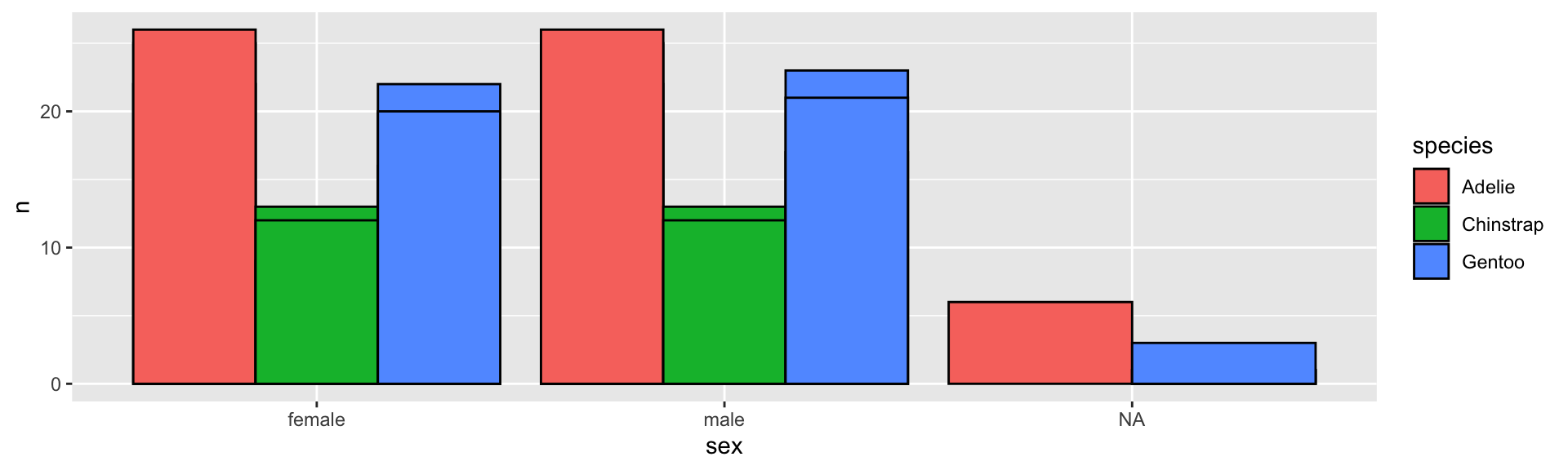
- Here the
xvalues are recalculated so that the factor levels within the same group (as determined byx) can fit.
Another grouped barplot with geom_col()
penguins %>%
group_by(sex, species, year) %>%
tally() %>%
ggplot(aes(sex, n, fill = species, group = year)) +
geom_col(color = "black", position = "dodge2")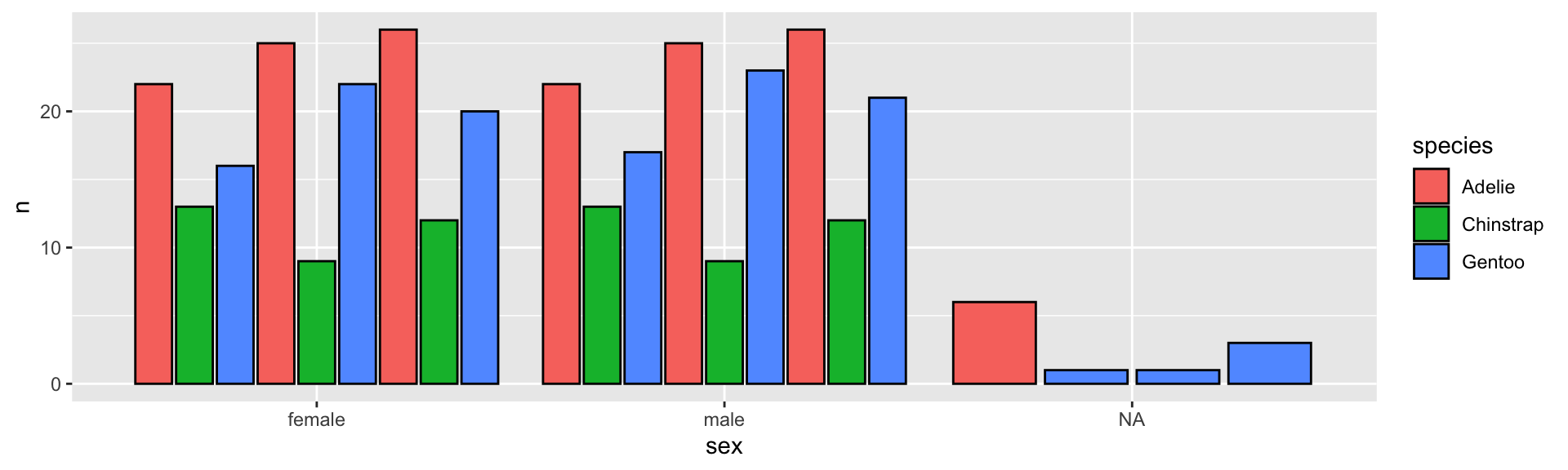
position = "dodge"doesn’t deal well when there isfillandgrouptogether but you can useposition = "dodge2"that recalculates thexvalues in another way.
Stacked percentage barplot with geom_col()
penguins %>%
group_by(species, sex, year) %>%
tally() %>%
ggplot(aes(sex, n, fill = species, group = year)) +
geom_col(color = "black", position = "fill")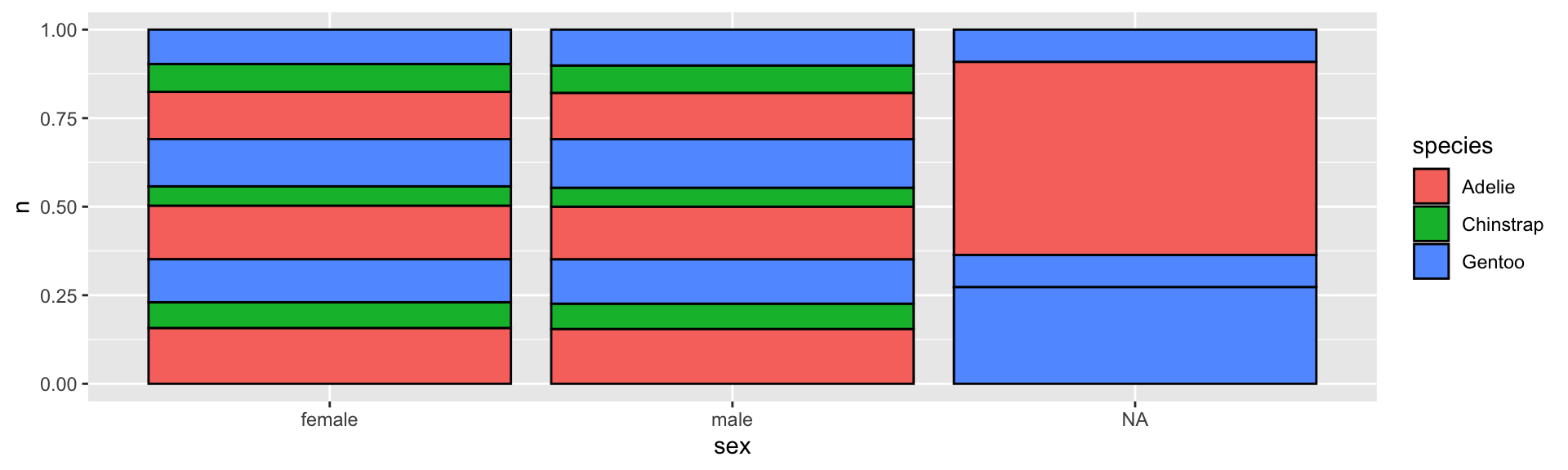
- If you want to compare the percentages between the different
x, thenposition = "fill"can be handy.
Coordinate systems
Pie or donut charts with coord_polar()
- The default coordinate system is the Cartesian coordinate system.
Other coordinate systems
coord_cartesian()for Cartesian coordinate systems (default)coord_flip()to flip the x and ycoord_fixed()to use a fixed aspect ratiocoord_equal()is essentiallycoord_fixed(ratio = 1)coord_trans()to transform the coordinate after the statistical transformationcoord_map()to use projection based onmapproj
Your turn!
10:00
> Go to emitanaka.org/dataviz-workshop/exercises/
> Click Exercise 4